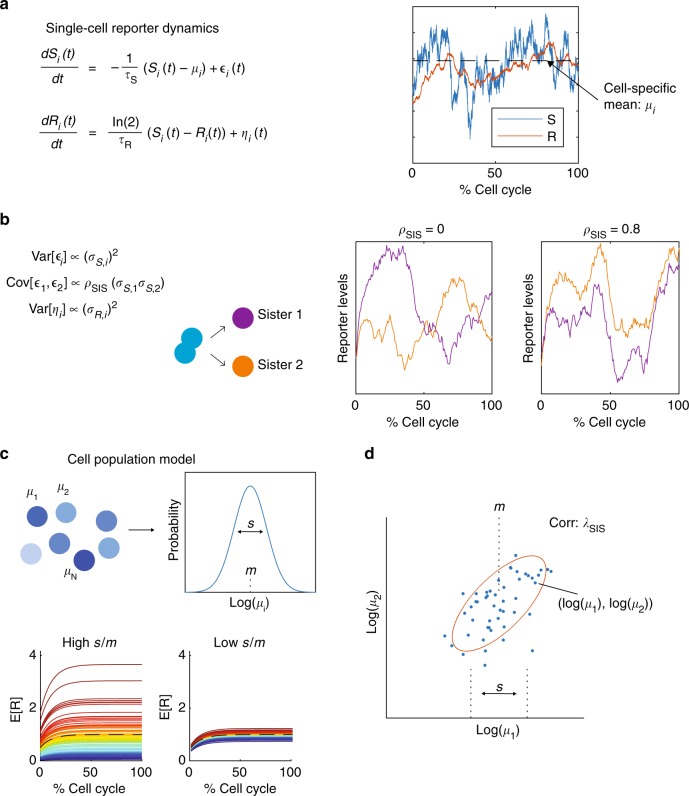
Fig. 3.
A hierarchical model of single-cell reporter dynamics in pairs of sister cells. a Single-cell dynamics are modelled probabilistically using stochastic differential equations (Methods). Each cell has a transcriptional activity (S) and a bioluminescent reporter (R) variable, where S controls the production of R. b To account for stochastic fluctuations, both S and R are perturbed by noise terms ε and η, respectively. The transcriptional noise experienced in the two sister cells is correlated with parameter ρSIS, which describes whether sister cell dynamics are independent (ρSIS = 0) or if they share a similar shape over the cell cycle (ρSIS > 0). c The mean level of S is cell-specific and denoted by μi for cell i, and the strength of the noise terms for S and R are also cell-specific and are denoted by σS,i and σR,i, respectively. The distribution of cell-specific parameters μi, σS,i and σR,i are described at the population level with log-normal distributions. s describes the population level variability in cell-specific means. d The correlation of mean transcriptional levels between sister cells is quantified with λSIS