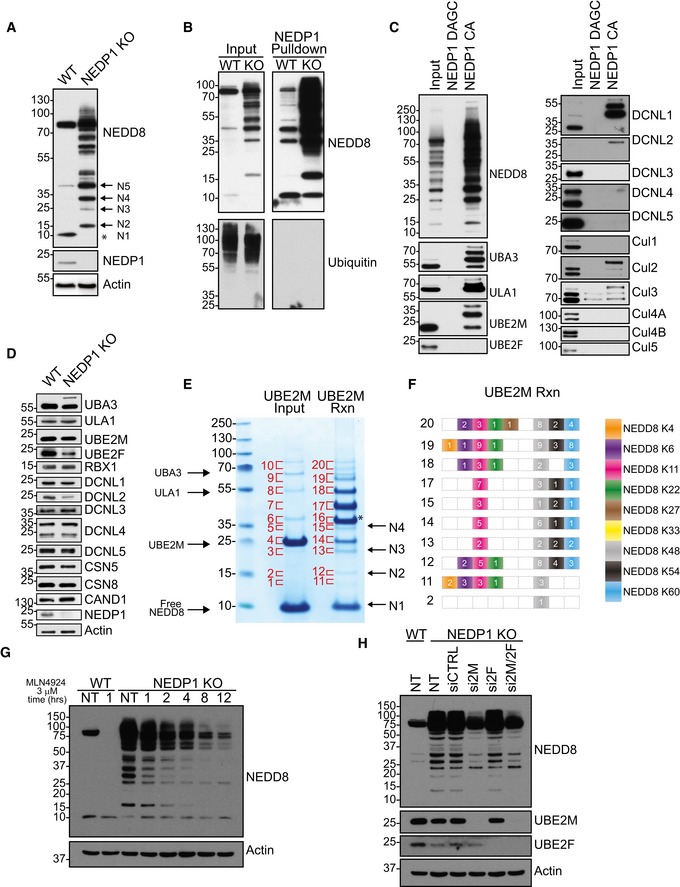
Western blot analysis of whole‐cell lysates from HEK 293 WT and NEDP1 KO cells reveals a loss of free NEDD8 (indicated by asterisk) and an accumulation of NEDD8 reactive species in the NEDP1 KO lysate. The predicted molecular weight sizes of putative, unanchored, poly‐NEDD8 chains are denoted by N2 through to N5. Unconjugated NEDD8 is denoted by N1.
NEDD8 affinity resin shows enrichment of endogenous neddylated proteins in WT and NEDP1 KO cells. Recombinant HALO‐NEDP1 C163A (CA) conjugated to HALO‐Link beads was used as an affinity resin to enrich for neddylated proteins in lysates from HEK 293 WT and NEDP1 KO cells. Enriched proteins were resolved by SDS–PAGE and processed for Western blot analysis with NEDD8 or ubiquitin antibodies. HALO‐NEDP1 CA specifically enriches for NEDD8‐reactive proteins in both WT and NEDP1 KO cells, but does not enrich for Ubiquitin‐modified proteins in either cell line.
Components of the NEDD8 conjugation machinery are enriched in HALO‐NEDP1 pulldowns from NEDP1 KO lysates. Neddylated proteins from HEK 293 KO cells were enriched by HALO‐NEDP1 CA pulldown, as in (B) but not by the NEDD8 nonbinder mutant, HALO‐NEDP1 DAGC (D29W A98K G99K C163A). The NEDD8 E1s, UBA3 and ULA1, are modified in NEDP1 KO cells, as well as E2 UBE2M, and co‐E3s DCNL1 and DCNL2. Cul2 and Cul3 are hyper‐neddylated in NEDP1 KO cells. CSN components, CSN5 and CSN8, also co‐precipitate in HALO‐NEDP1 CA pulldowns.
Western blot analysis from HEK 293 WT and NEDP1 KO cells of the components of the NEDD8 conjugation/de‐conjugation pathway shows that similar levels of NEDD8 pathway components are present in both WT and NEDP1 KO cells. Apart from UBA3, there is no detectable amount of NEDD8‐modified enzymes in whole‐cell lysates from NEDP1 KO cells.
Poly‐NEDD8 chains can be generated by in vitro reactions (Rxn). NAE (0.15 μM), UBE2M and NEDD8 (20 μM) were incubated on ice or incubated at 30°C for 3 h and reactions were stopped by addition of LDS sample loading buffer. Reactions were resolved by SDS–PAGE and stained with colloidal Coomassie. Indicated bands were excised from the gel and processed for in‐gel trypsin digestion and mass spectrometry analysis. The predicted molecular weight sizes for a theoretical unanchored NEDD8 chain are denoted by N2‐N4. Unconjugated NEDD8 is indicated by N1. UBE2M modified by NEDD8 is indicated with an asterisk.
Diagram of the NEDD8 linkages, as determined by mass spectrometry analysis, from (E), with the number of spectral counts indicated for the bands labelled in (E). Only bands with identified diGly motifs are shown here. UBE2M generates in vitro chains of poly‐NEDD8 with linkages on K4, K6, K11, K22, K27, K48, K54 and K60.
Neddylated species are NEDD8 E1 dependent. WT and NEDP1 KO HEK 293 cells were treated with NAE inhibitor MLN4924 at 3 μM for the indicated time. Lysed cells were then processed for Western blot analysis. NEDD8 E1 inhibition results in a time‐dependent decrease in the amount of Cullin and non‐Cullin NEDD8 reactive bands.
Neddylated species are UBE2M dependent. WT and NEDP1 KO HEK 293 cells were left untreated or treated with the indicated siRNA for 48 h. Lysed cells were then processed for Western blot analysis. Neddylated species are reduced when NEDD8 E2 UBE2M was depleted after siRNA treatment (si2M), but not with control siRNA (siCTRL) or when NEDD8 E2 UBE2F was depleted (si2F). The double knockdown (si2M/2F) does not further reduce NEDD8 modified proteins.