Figure EV1. Analysis of the RsaI expression.
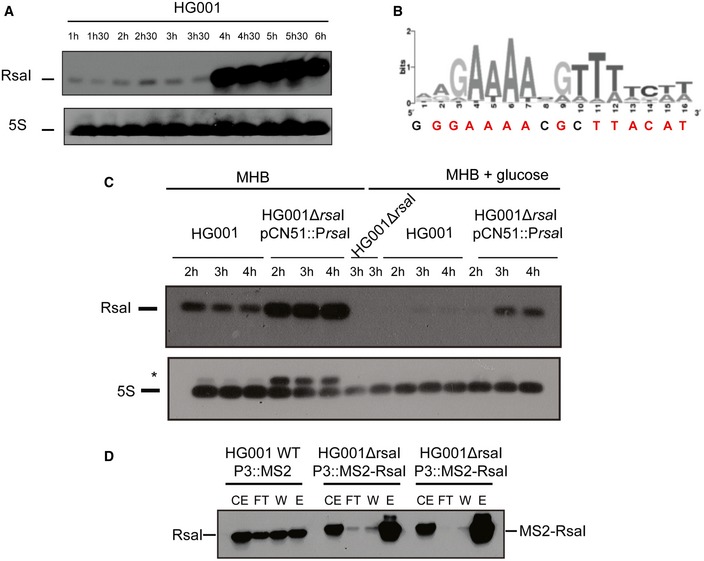
- Northern blot experiments showing the expression of RsaI in HG001 strain during growth phase. Total RNA was prepared from samples taken after various time points of growth in BHI medium at 37°C. Hybridization against 5S rRNA was used as loading control on the same samples which have been migrated on another agarose gel because RsaI and 5S rRNA have very similar sizes.
- The cre‐site consensus was defined in Bacillus cereus (van der Voort et al, 2008). The sequence found in the 5′ region of RsaI is represented below the graph, the conserved residues are in red.
- Northern blot experiments showing the expression of RsaI in HG001 strain, the isogenic HG001∆rsaI mutant strain, and the same mutant strain complemented with a plasmid expressing RsaI under the control of its own promoter (HG001∆rsaI pCN51::PrsaI). Total RNA was prepared from samples taken after various time points of growth in MHB medium in the absence or presence of 1% glucose. For the mutant HG001∆rsaI strain (used as a negative control), total RNA was prepared after 4 h of growth in MHB medium in the absence or in the presence of glucose. Hybridization against 5S rRNA was used as loading control on the same membrane. *Traces of RsaI signal after re‐hybridization of the membrane with the 5S probe were still observed.
- MS2‐RsaI is specifically retained by affinity chromatography containing the MS2‐MBP protein. The Northern blot was performed using a DIG‐labeled RsaI probe (Table EV3) to visualize RsaI and MS2‐RsaI following the MS2 chromatography affinity. CE is for crude extract, FT for flow‐through, W for washing, and E for elution. For CE/FT/W samples, 5 μg of total RNA was loaded on a 1.5% agarose gel while for E sample, only 0.5 μg of total RNA was used. The two replicates are shown for the RNA purified from the strain HG001ΔrsaI::MS2‐RsaI.
Source data are available online for this figure.
