Figure 2. RsaI binds to icaR, glcU_2, fn3K mRNA, and the sRNA RsaG.
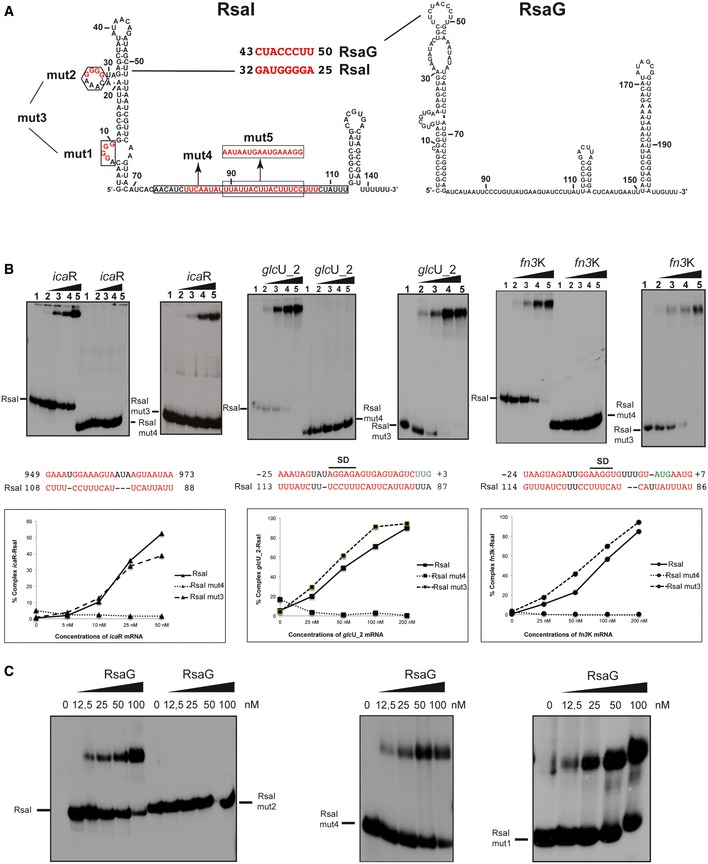
- Secondary structure model of RsaI. In red, are the nucleotides deleted in the RsaI mutants (mut1 to mut4), and the nucleotides, which were substituted (mut5). The potential base‐pairings between RsaI and RsaG are shown. Squared nucleotides are conserved sequences in RsaI.
- Gel retardation assays show the formation of the complex between RsaI and icaR, glcU_2 and fn3K mRNAs. The 5′ end‐labeled wild‐type RsaI (RsaI), RsaI mutant 3 (RsaI mut3, deletion of the two G‐track motifs), and RsaI mutant 4 (RsaI mut4, deletion of the C/U‐rich unpaired interhelical region) were incubated with increasing concentrations of mRNAs: lane 1, 0 nM; lane 2, 25 nM; lane 3, 50 nM; lane 4, 100 nM; and lane 5, 200 nM. Below the gels, the predicted interactions between RsaI and its targets are shown. Translation start codons are in green, and SD is for Shine and Dalgarno sequence. Graphs represented the % of complex formed between either RsaI or its two mutant forms (RsaI mut3 and RsaI mut4) and the target mRNA (icaR, glcU_2, fn3K) as the function of mRNA concentrations.
- Gel retardation assays show the formation of the complex between RsaI and RsaG. The 5′ end‐labeled wild‐type RsaI (RsaI), RsaI mutant 2 (RsaI mut2), RsaI mutant 4 (RsaI mut4), and RsaI mutant 1 (RsaI mut1) were incubated with increasing concentrations of RsaG given in nM on the top of the autoradiographies.
Source data are available online for this figure.
