Figure 4. Interaction of RsaI to icaR mRNA induces biofilm production.
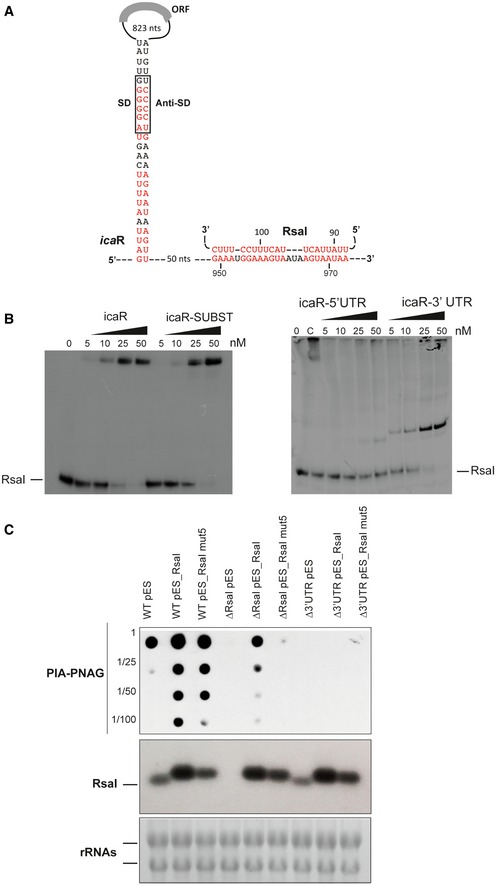
- Secondary structure model of icaR mRNA. The nucleotides in red are involved in the long‐range interaction between the 3′ and 5′UTRs or with RsaI. SD and anti‐SD are, respectively, for the Shine and Dalgarno sequence and for the sequence complementary to the SD sequence.
- Gel retardation assays show the formation of the complex between RsaI and icaR full‐length, icaR SUBST, icaR‐5′UTR, and icaR‐3′UTR. The 5′ end‐labeled of RsaI was incubated with increasing concentrations of the various mRNAs. UTR is for untranslated region, and SUBST stands for the substitution of UCCCCUG sequence by AGGGGAC. This mutation, which is located in the 3′UTR of icaR, significantly destabilizes the long‐range interaction and enhances icaR translation. Lane C represents binding between radiolabelled RsaI and full‐length icaR mRNA (50 nM). Due to the rather short migration of the gel, the full‐length mRNA is still observed in the pocket.
- In vivo effect of RsaI on PIA‐PNAG synthesis in the S. aureus wild‐type (WT) 132 strain, the ΔrsaI mutant, and the strain carrying a deletion of icaR 3′UTR (Δ3′UTR). This last mutant strain has been transformed with the pES plasmids expressing rsaI or rsaI mut5. The PIA‐PNAG exopolysaccharide biosynthesis was quantified using dot‐blot assays. Serial dilutions (1/5) of the samples were spotted onto nitrocellulose membranes, and PIA‐PNAG production was detected with specific anti‐PIA‐PNAG antibodies. RsaI was detected in the same samples by Northern blot using a probe directed against RsaI. Ethidium bromide staining of rRNA was used as loading controls of the same gel.
Source data are available online for this figure.
