Abstract
Accurate transmission of the genetic information requires complete duplication of the chromosomal DNA each cell division cycle. However, the idea that replication forks would form at origins of DNA replication and proceed without impairment to copy the chromosomes has proven naive. It is now clear that replication forks stall frequently as a result of encounters between the replication machinery and template damage, slow-moving or paused transcription complexes, unrelieved positive superhelical tension, covalent protein-DNA complexes, and as a result of cellular stress responses. These stalled forks are a major source of genome instability. The cell has developed many strategies for ensuring that these obstructions to DNA replication do not result in loss of genetic information, including: DNA damage tolerance mechanisms such as lesion skipping, where the replisome jumps the lesion and continues downstream, template switching both behind template damage and at the stalled fork, and the error-prone pathway of trans-lesion synthesis.
Keywords: DNA lesion, replication fork stalling, lesion skipping, trans-lesion synthesis, replication fork reversal, template switching, replication restart
1. INTRODUCTION
The enzymatic pathways that act to maintain continued replication in the face of replication conflicts, that preserve the integrity of the stalled fork and of the replication machinery, that remodel and repair the stalled forks, and that restart replication when it is arrested are crucial to survival of the organism. In humans, defects in the genes encoding some proteins involved in the repair of damaged replication forks cause chromosome instability syndromes and confer increased risk of various cancers (1–3). The importance of understanding the how and why of fork stalling, lesion bypass, and the consequences of failed replication restart is illustrated by the overwhelming number of excellent reviews that have appeared in the last few years [for a sampling see (4–11)]. Many of these reviews are comprehensive and highly detailed in their analyses of different aspects of the topic. This review will focus more on the mechanisms underlying events at stalled forks and discuss the impact of some recent data on our understanding of these events.
2. REPLISOMES
The antiparallel nature of the two strands of DNA in the double helix and the direction of polymerization of all DNA polymerases dictates a difference in the nature of synthesis of the two nascent strands. The nascent leading strand can be synthesized continuously in the 5′ → 3′ direction (although see Sections 3.1 and 3.2 below), whereas the nascent lagging strand is synthesized in short, discontinuous fragments (Okazaki fragments) that are subsequently joined together after the replication fork has proceeded down the template (12).
The assembly of proteins that is responsible for DNA replication is often called the replisome, implying physical association of the individual components to form a multi-protein complex. It is important to distinguish between the replisome and a replication fork, the latter being the point on the template DNA at which the duplex DNA is being unwound and nascent strand synthesis is occurring. The terms are, unfortunately, often used interchangeably, with “replication fork” used to describe the action of a replisome at a replication fork. But when replication fork progression—the consequence of replisome action at a replication fork—is stalled by template damage, what happens to the replisome and the replication fork are often quite different.
Four key activities are required to sustain replication fork progression. These are: (i) the leading- and lagging-strand polymerases that synthesize the nascent DNA, (ii) a DNA helicase to unwind the parental duplex strands prior to copying by the DNA polymerases, (iii) a primase to synthesize short oligoribonucleotide primers for the initiation of synthesis on the lagging-strand template (as well as occasionally on the leading-strand template), and (iv) a single-stranded DNA-binding protein to protect exposed single-stranded (ss) DNA from degradation, eliminate secondary structure in the template, and serve as a platform for binding of other proteins involved in replication and repair. Eukaryotic cells require additional activities to displace nucleosomes ahead of the advancing fork and re-establish them behind it.
The prokaryotic replisome is relatively simple compared to the eukaryotic one. The leading- and lagging-strand polymerases are identical and provided by the multi-subunit DNA Polymerase III Holoenzyme (DNA Pol III HE) (13). The Pol III HE also includes the DnaX complex that holds the polymerase subunits together via interactions between the τ subunit of the DnaX complex and the catalytic polymerase α subunits. This complex also serves as the loader of the DNA polymerase topological processivity clamp β. The DNA helicase is the hexameric DnaB that travels 5′ → 3′ on the lagging-strand template while it unwinds the DNA. The primase is DnaG, which transiently associates with DnaB to synthesize primers. DnaB also interacts with τ, cementing the association between the three principal catalytic activities of the replisome. And the single-stranded DNA-binding protein is SSB, which also interacts with subunits of the DnaX complex and DnaG.
The bacterial replisome is loaded at the single bacterial origin, oriC, in a sequence-specific manner that involves the interaction of the initiator protein DnaA with DnaC, a chaperone for DnaB, as well as with DnaB itself (14). The presence of only one origin on most bacterial genomes renders fork progression-blocking events lethal in the absence of origin-independent replication reactivation functions.
Recent seminal studies reporting the reconstitution of yeast DNA replication with purified proteins have given us a clear view of the components of the minimal eukaryotic replisome and associated activities required to sustain replication fork progression (15–19). Three DNA polymerases act at the eukaryotic replication fork. Pol α/primase initiates synthesis of both the nascent leading and lagging strands. Pol δ synthesizes the nascent lagging strand [and can synthesize the nascent leading strand under certain conditions (17, 19)], whereas Pol ε synthesizes the nascent leading strand [although Pol δ acts to extend the Pol α/primase primers somewhat before Pol ε engages (17)], suggesting frequent trafficking of DNA polymerases at the 3′-end of the nascent leading strand. The processivity polymerase clamp is PCNA, which has differential effects on the three DNA polymerases: Little stimulation of Pol α, strong stimulation of Pol δ, and moderate stimulation of Pol ε, which, even in the absence of PCNA, is fairly processive (20). Nevertheless, maximally efficient DNA replication in vitro requires the action of PCNA on the nascent leading strand (17, 19). The five-subunit RFC complex (Rfc1–5) is responsible for loading PCNA onto DNA.
The core and motor of the eukaryotic DNA helicase is the hexameric Mcm2–7 complex. A double hexamer of Mcm proteins is loaded in inactive form to the DNA by ORC, Cdc6, and Cdt1 during G1 phase. Activation of the helicase activity during S phase requires the protein kinase activities of DDK and CDK and the action of Sld2, Sld3, Sld7, Dpb11 (none of which are thought to be associated with the activated helicase), Cdc45, GINS, Mcm10, RPA—the eukaryotic SSB—and Pol ε. The complete, activated DNA helicase is called the CMG complex and comprises, in addition to the Mcm proteins, the GINS complex and Cdc45. Unlike DnaB, the CMG complex travels 3′ → 5′ on the leading-strand template. Interestingly, recent cryo-EM data (21) demonstrated that CMG travels with its C-terminal DNA helicase domain forward and the N-terminal domain behind, the opposite of DnaB, indicating that upon activation, the two Mcm hexamers have to pass each other, as has been proposed for replisome establishment at oriC (22).
Maximal rates of replication in vitro, equivalent to those found in vivo, required several components of the replication fork protection complex (23): Ctf4, Mrc1—which may stimulate CMG-catalyzed unwinding directly—and the Csm3/Tof1 complex, which is thought to stabilize the association of Mrc1 with the replisome components Mcm and Pol ε (17). Replication fork progression through nucleosome-coated templates further required the histone chaperone, FACT and its associated protein Nhp6, the lysine acetyl transferases Gcn5 and Esa1, as well as either the INO80 or ISW1A nucleosome remodeling complexes (18, 19).
Note that loading to the DNA template of the replisomes described above occurs in an origin-dependent manner, requiring regulated processes and specific DNA-binding proteins. Such processes are not available to reactivate replication fork progression that has been stalled because of collision with an obstruction. Various strategies that have evolved for cells to overcome this problem are discussed below.
3. STALLING OF REPLICATION FORK PROGRESSION, REMODELING OF STALLED FORKS, AND REPLICATION REACTIVATION
The encounter of replisomes with damage in the leading- and lagging-strand templates have different consequences. As long as unwinding by the helicase is not impaired, damage in the lagging-strand template will not hamper replication fork progression in any significant way. This is because replication on that strand is already geared to constantly initiate new Okazaki fragments. Thus, a lagging-strand polymerase that does become stalled at a template lesion will be able to cycle forward rather soon to the next primer synthesized. Alternatively, the lagging-strand polymerase may remain stalled at the lesion, dissociate from the replisome, and be replaced by another polymerase from the soluble store as the replisome continues to move forward. In support of the view that template damage is not a challenge to lagging-strand replication, replication fork progression with the E. coli proteins in vitro is unimpeded by an abasic site in the lagging-strand template (24). Therefore the net effect of lagging-strand template damage is that gaps are left behind in the nascent lagging strand that have template damage directly opposite the 3′-end of the gap (similar to the case in Fig. 1e, although the gap shown there is in the nascent leading strand).
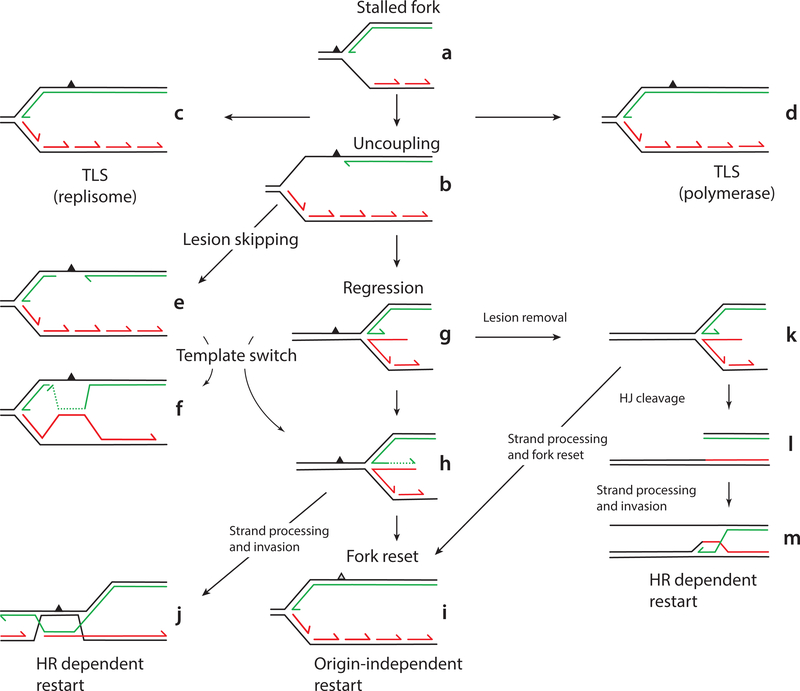
Figure 1.
Pathways of lesion bypass and replication fork reactivation. (a) a replication fork progressing from right to left that has just been stalled by a leading-strand template lesion (the black triangle). Template strands are black, nascent leading strands are green, and nascent lagging strands are red. See text for explanations of the pathways. Direct TLS by the replisome (a to c). Polymerase switching from the replicative polymerase to a specialized TLS polymerase and back to the replicative polymerase (a to d). Functional uncoupling of leading-strand DNA synthesis from template unwinding and lagging-strand synthesis (a to b). Lesion skipping (b to e). Post-replicative template switching (e to f). Replication fork reversal (b to g). Template switching in a reversed fork (g to h). Fork reset, the reversed fork is restored to the original configuration of nascent and template strands (h to i). HR-dependent replication restart for a reversed fork (h to j). Lesion removal by nucleotide excision repair at a reversed fork (g to k). Cleavage of a reversed fork by Holliday junction resolvases (k to l). HR-dependent replication restart after Holliday junction cleavage (l to m). Note that the reversed fork in (k) can also be reset directly (k to i), with the grayed out damage triangle indicating that in this pathway, the template damage would have been removed prior to fork reset. Replication at the reset fork (i) can be restarted by origin-independent replisome loading activities.
Damage in the leading-strand template is more problematic and it is there we will focus to describe the many pathways reported to act to prevent genome instability at a stalled fork (Fig. 1). When a replisome first stalls at leading-strand template damage, the situation drawn in Fig. 1a obtains. At this juncture the nascent lagging strand has not yet caught up with the stalled leading strand. Subsequently, it is typically posited that “uncoupling” of leading- and lagging-strand synthesis occurs, that is, leading-strand synthesis is halted and lagging-strand synthesis and template unwinding continues (Fig. 1b) (25–27). This event generates ssDNA as the leading-strand template is exposed that becomes coated with either SSB or RPA. In bacteria, the SSB-coated ssDNA is the trigger that induces the SOS response (28); whereas in yeast (29, 30) and mammalian cells (31, 32), the RPA-coated ssDNA induces the S-phase checkpoint.
3.1. Uncoupling of Leading- and Lagging-strand Synthesis
Studies using the E. coli replication proteins and a template carrying a single cyclopyrimidine dimer (CPD) in the leading-strand template supported the concept of polymerase uncoupling (33). It was observed that when the leading-strand polymerase stalled at the lesion, DnaB continued unwinding the template at a reduced rate (35–50 nt/s) as lagging-strand synthesis continued, a classic description of polymerase uncoupling. However, we now appreciate that this behavior is simply a manifestation of the intrinsic lack of coordination of synthesis by the two polymerases Real-time single-molecule studies of rolling-circle DNA replication with E. coli proteins (34) has given us some unexpected findings. Many models of DNA replication posit that synthesis of the leading- and lagging-strands are coordinated so that large ss gaps do not appear in the nascent DNA. Because there are several additional time-requiring steps during the synthesis of an Okazaki fragment compared to leading-strand synthesis (recognition of a priming signal by the primase, primer synthesis, termination of synthesis of the penultimate Okazaki fragment, formation of a new DNA polymerase-processivity clamp complex on the new primer terminus, and commencement of lagging-strand synthesis) it has been proposed that the lagging-strand polymerase had to synthesize DNA at a faster rate than the leading-strand polymerase [see, for example, (35)] or that priming might transiently halt leading-strand synthesis (36).
Graham et al. (34), however, observed that overall replication fork progression was unaffected by whether lagging-strand synthesis was ongoing. Furthermore, the average rates of DNA synthesis by the leading- and lagging-strand polymerases were similar and the rate of polymerization by either polymerase could vary by a factor of ten. Remarkably, individual trajectories of active leading- and lagging-strand polymerases showed distinct pauses with the rate of polymerization switching stochastically after a pause. DNA unwinding continued during these pauses but did so at 20% of the average rate of fork progression. This reduced rate of unwinding while the DNA polymerase is paused prevents the accumulation of unreplicated gaps in the DNA. It is thus a fail-safe mechanism or “dead-man’s switch,” that prevents the DNA helicase from running away from the polymerases. The dead-man’s switch permits functional recoupling of the helicase and polymerase and resumption of fork progression at typical rates.
The sampling of wide distributions of rates and stochastic polymerase pausing obviates the need to impose differential rates for “coordinated” replication. This fluctuation in rates solves the coordination problem—there is no coordination. Constant pausing and switching of polymerization rates by the polymerases implies that at any given time the leading-strand polymerase could be synthesizing DNA faster or slower than the lagging-strand polymerase and vice-versa. Whereas transient gaps may thus form on any DNA template, they will be filled in over a short time range as the rates of synthesis vary. These observations imply that the replication process is more dynamic than previously thought, with physical contacts within the replisome likely to be continually broken and reformed. Seen in this light, so-called “uncoupling” of the leading- and lagging-strand polymerases is simply the net effect of the dead-man’s switch in action and is not derived per se from the encounter of the leading-strand polymerase with template damage.
This stochastic model of DNA replication makes it more likely that a replisome can progress past template damage: If pauses and recovery are inherent to replisome action, then pausing at damage is part of normal replisome function. Indeed, such pausing may have been built in evolutionarily to accommodate template damage. Can we extend this model to the eukaryotic replisome? No similar single-molecule analyses with the eukaryotic replisome have been reported as yet, although the recent advances in reconstituting the replication system suggests that we will soon see such studies. However, there are some features of the eukaryotic replisome that could be seen as supportive of this model.
The eukaryotic replisome, as the prokaryotic one, is typically drawn as a tight complex of many different proteins [see, for example, (20)], and, indeed, the eukaryotic replisome possesses associated proteins tasked with maintaining the stability of the complex (23). Furthermore, cryoelectron microscopy and biochemical studies reveals a tightly associated complex of CMG and pol ε (21, 37–39), but this picture doesn’t really speak to whether the polymerases can act independently. The Ctf4 trimer may be the functional equivalent of the τ subunit of the DNA Pol III HE, interacting with Pol α, GINS, and Pol ε (40, 41). Furthermore, the C-terminal, noncatalytic domain of the POL2 subunit of Pol ε interacts directly with the CMG, whereas the catalytic N-terminal domain is relatively unconstrained in space (21, 39). On the other hand, no interaction between Pol δ and CMG could be detected (42), thus this polymerase seems to find its way to the replisome by its affinity for PCNA, possibly accounting for its ability to synthesize both nascent leading and lagging strands in the reconstituted yeast system under certain conditions (16–19).
Pol α and Pol δ clearly have access to both template strands, whereas the cryo-EM structures suggest that Pol ε may be restricted to the leading-strand template. Schauer and O’Donnell (42) have suggested that any Pol δ-PCNA complex on the leading-strand template will catch up with the slow-moving CMG complex and be dissociated by collision release, whereas involvement of Pol ε on the lagging-strand template is inhibited by RFC. Thus, the distribution of any particular polymerase activity in the eukaryotic replisome could well be independent of the action of the others.
3.2. Lesion Skipping
Early observations in E. coli, yeast, and human cells suggested that after UV irradiation, the nascent DNA is discontinuous on both strands and gaps remain opposite the lesions formed (43–45). To account for this phenomenon, Rupp and Howard-Flanders (45) proposed that replication could be re-initiated downstream from lesions in both the leading- and lagging-strand templates. Forty-three years later a mechanistic basis was provided to support this model (46). This study used the E. coli replication proteins and a template with a specifically placed CPD in the leading-strand template and demonstrated that whereas the damage did transiently stall the replisome, the replisome remained associated with the DNA template and leading-strand synthesis could resume when the stalled leading-strand polymerase cycled forward past the lesion to a new primer made downstream on the leading-strand template by DnaG. As described in the previous section, this template strand was made available by the continued unwinding of the parental duplex by the DnaB helicase. Thus, at least the E. coli replisome has the inherent capacity to replicate beyond leading-strand template damage by synthesizing the nascent leading-strand discontinuously. This DNA damage tolerance reaction has been termed ‘lesion skipping’ (follow Fig. 1a to 1b to 1e). In retrospect, such a mechanism is a direct prediction of the stochastic model of DNA replication (34) described above. The damage left behind in the gap can then be repaired either by homology-directed gap repair processes or the action of a trans-lesion synthesis (TLS) DNA polymerase followed by gap filling.
Does a similar reaction occur in eukaryotic cells? Lopes et al. (27), using two-dimensional gels and electron microscopy to examine newly replicated DNA after UV-irradiation in yeast cells, reported that replication slowed, but did not stop, and that small DNA gaps could be observed in the replicated duplexes. Similar gaps have been reported in S. pombe (47, 48). And, Elvers et al. (49) found, using DNA fiber analysis after UV-irradiation in human fibroblasts, that replication fork progression was only slightly reduced and concluded that repriming downstream of the lesions was likely occurring.
The lesion skipping reaction requires both a repriming event downstream of the lesion and the continuance of unwinding of the template strands. Fumasoni et al. (50) demonstrated that in yeast primase mutants (pri1) challenged with the alkylating agent methyl methanesulfonate, template-switching—post-replicative, gap-repair pathways behind the replication fork (Fig. 1b to 1e to 1f, and see below)—were disfavored and fork remodeling pathways (Fig. 1b to 1g) became favored. They concluded that Polα/primase priming downstream of the lesions was therefore normally operative.
Repriming downstream of lesions in mammalian cells is accomplished by PrimPol, the second member of the Archaeo-Eukaryotic Primase family to be found in human cells (51). This interesting enzyme has DNA polymerase, TLS, and primase activities, the latter of which can synthesize a primer from only dNTPs (it also uses NTPs) (52–55). Even more interesting is that the primase activity of this enzyme is required for tolerance of UV-irradiation in human (56) and chicken DT-40 (57) cells, as well as tolerance to DNA damage induced by treatment with methyl methanesulfonate and cisplatin in the latter cell type.
A plausible pathway for lesion skipping in yeast and mammalian cells could therefore be as follows for leading-strand template damage (Fig. 1b to 1e): Pol ε stalls at the template damage whereas the CMG continues to unwind the parental template duplex and Pol α/primase and Pol δ continue to synthesize the nascent lagging-strand. Because its affinity for PCNA is quite low [4% that of Pol δ (42)], Pol ε probably can dissociate quite readily from PCNA, or, alternatively, actually be dissociated or displaced by either RFC or Pol δ (42). However, given that the C-terminal domain of Pol ε will still be bound to the CMG, the polymerase will move away from the lesion along with the progressing helicase complex. At this point, Pol α/primase in yeast or PrimPol in mammalian cells can prime downstream of the damage on the leading-strand template. Pol δ is known to be able to readily detect 3′-ends and extend from them. For example, Pol δ is required for break-induced replication (BIR) (58), broken fork repair (59), and homologous recombination (HR)-dependent fork restart (see below) (60). Thus, Pol δ can extend from the new primer and catch up with the progressing CMG-Pol ε complex, dissociating by collision release when it collides with the slow-moving CMG complex (42). Such a role for Pol δ was recently proposed by Yeeles et al. (17). Of course, an unknown here is the role of the fork protection proteins Mrc1, and the Csm3/Tof1 complex [Claspin, Tipin, and Tim (Timeless) in metazoans, respectively]. The activity of these proteins seems situational: In yeast they mediate the S-phase checkpoint at stalled forks (61, 62), yet, as described above, they are also required for maximal rates of replication fork progression in the reconstituted system (17). How lesion skipping fits into this framework and how all these potential reactions at stalled forks are regulated remains to be determined.
3.3. Trans-lesion Synthesis
It has been accepted that replicative DNA polymerases, those that act in the replisome to synthesize DNA, are intolerant of DNA template lesions. X-ray crystal structures of TLS and cellular replicase polymerases have revealed a striking difference in configuration of the active sites. Cellular replicases tend to be tightly apportioned, with no “room” in the active site to accommodate either non-standard base conformations or rotation of template bases out of line with the template duplex, whereas the active sites of TLS polymerases are far “roomier,” able to tolerate such non-standard deviations (63, 64).
Three E. coli DNA polymerases have TLS activity, Pol II, Pol IV [DinB (65)], and Pol V [UmuD′2UmuC (66, 67)]. There are many TLS polymerases in yeast and mammalian cells, the major ones being Pol η, Pol ζ (a tetramer of Rev3, Rev7, and the Pol31 and Pol 32 subunits of Pol δ), Rev1, Pol ι, and Pol κ (a DinB homolog), the latter two being specific to mammalian cells. That these TLS DNA polymerases are required to bypass template damage to maintain genomic integrity is clear. In E. coli umuC and umuD mutants are very devoid of most UV-induced mutagenesis (68, 69) and in humans Pol η is encoded by the xeroderma pigmentosum variant (XP-V) locus. The different TLS polymerases have selective activities on different types of template damage and, at least in mammalian cells, appear to be divided into bypass polymerases and those that extend from the base inserted opposite the damage (70–73)
An alternative to the lesion skipping reaction is the recruitment of specialized DNA polymerases that can switch with the stalled replicative polymerase, bypass the DNA damage by TLS, and then allow the replicative polymerase to resume replication (Fig. 1a to 1d). The original idea for the polymerase switching model, the ‘toolbelt model” [formulated initially based on observations with β (74), but described here for both β and PCNA] arose from the demonstration that the processivity clamps, β and PCNA, were toroidal multimers (a dimer and trimer, respectively) and that various DNA polymerases shared similar amino acid sequences for interacting with the clamps [β binding domain; PCNA interacting peptide (PIP)]. These mostly α-helical peptides, which are generally at either the N- or C-terminus of the polymerases, interact with inter-domain connecting loops on the clamps. Thus, if the replicative polymerase was engaged with one of the subunit of the clamps, other TLS DNA polymerases could potentially be engaged with another subunit and be readily available in place to switch with the replicative polymerase at a lesion, possibly by rotation of the loaded clamp.
In support of the toolbelt model, Indiani et al. (74) demonstrated, using complementary strand synthesis on a primed M13 ssDNA template as an assay, that Pol IV could switch with a stalled Pol III, but not one that was moving. These same authors demonstrated (75), using a rolling circle DNA replication system, that both E. coli Pol II and Pol IV could support slow DNA replication with the DnaB helicase and could remodel a moving Pol III HE replisome by slowing it down. And using single molecule analysis, Kath et al. (76) demonstrated exchange between E. coli Pol IV and the Pol III HE during primer extension on a ssDNA template. Similarly, McCulloch et al. (77) demonstrated the sequential action of Pol δ and Pol ε with Pol η during bypass of a CPD using a primer extension assay.
Is it necessary to invoke simultaneous occupancy of one clamp molecule by multiple DNA polymerases to achieve switching? The observations outlined above could also be explained by one polymerase dissociating from the clamp because of local conditions and the next polymerase associating with the clamp left behind [see (8) for other arguments along these lines] Indeed, a cryo-EM structure of a presumed elongating complex of the Pol III HE with DNA shows that both of the hydrophobic inter-domain connecting loops on the β dimer are occupied: one by the α catalytic polymerase subunit and the other by the ε proofreading exonuclease subunit (78). Furthermore, recent single molecule studies in vivo and in vitro argue against a monolithic, stable replisome structure as depicted in many cartoon models, and argue for very rapid exchange of DNA polymerases during DNA replication.
The observations (34) that led to the stochastic model of DNA replication described above demonstrated that both the leading- and lagging-strand polymerases paused for considerable periods (a median of 13 s on the nascent leading strand). In those experiments replisomes were pre-formed onto rolling circle templates using the E. coli proteins and replication was observed in flow in the absence of additional DNA Pol III HE. In similar experiments that included the Pol III HE in the flow, Lewis et al. (79) demonstrated frequent and rapid exchange of the polymerases in the replisome dependent on the concentration of Pol III HE in the flow. Polymerase exchange, as demonstrated previously by Yuan et al. (80), was of the Pol III* particle containing the polymerase cores and the DnaX clamp-loading complex. At the highest concentrations of Pol III HE, exchange time was a few seconds. These authors also used cross-correlation analysis of fluctuating signals from two differentially fluorescently-tagged components of the Pol III HE to show that rapid exchange also occurred in vivo (exchange time of 4 ± 2 s). Similarly, Beattie et al. (81), using fluorescence recovery after photobleaching of fluorescently-tagged Pol III HE subunits and single-particle tracking Photoactivated Localization Microscopy, demonstrated that Pol III* subunits typically remained bound in the replisome for about 10 s. Notably, residence time of β was about 5-fold greater, whereas that of DnaB was 90-fold greater. Thus, at least in E. coli, rapid exchange of DNA polymerase subunits obviates the need for simultaneous occupancy of β, but clearly does point to β as being the platform for switching. Similar experiments as those described above for the E. coli system have yet to be reported with the eukaryotic system.
Other data arguing for the clamp, in this case PCNA, being the platform for trafficking of TLS polymerases of course comes from many studies on the role of ubiquitylation and SUMOylation of PCNA in modulating error-prone and error-free bypass pathways at stalled forks (72, 73, 82, 83). RPA-coated ssDNA that forms at stalled forks attracts the Rad18-Rad6 ubiquitin E3 and E2 enzymes that monoubiquitylate Lys 164 of PCNA. This event promotes the association of TLS polymerases with PCNA, facilitating the error-prone damage bypass pathway (Fig. 1a to 1d for bypass at the stalled fork or possibly Fig. 1a to 1b to 1e, for post-replicative bypass where the gap left behind after lesion skiping is filled in after bypass of the template damage by a TLS polymerase). Subsequent polyubiquitylation of PCNA on Lys164 by the E2 complex Ubc13-Mms2 and E3 enzyme Rad5 is thought to promote HR-dependent error-free pathways of bypass that involve template switches (see below).
The original model was that monoubiquitylated PCNA served to attract TLS polymerases to the stalled fork by direct binding, and, indeed, most eukaryotic TLS polymerases have both PIPs and ubiquitin binding domains (84, 85). With time, however, it became clear that TLS polymerase recruitment to stalled forks did not require PCNA ubiquitylation [see (73) for a summary and (86) for a recent biochemical analysis of Pol η binding to PCNA]. Attention has focused on the ability of Rev1, which interacts with PCNA through its N-terminal BRCT domain (87, 88) and polymerase-associated domain (89), to act as a scaffold protein. Boehm et al. (90) have demonstrated that the PIP motif of Pol η mediates its interaction with the C-terminal domain of Rev1. And single molecule analyses by the same group of protein complexes formed between PCNA, Rev1 and Pol η demonstrated that a PCNA toolbelt (where both polymerases bind to PCNA) and a Rev1 bridge (where both PCNA and Pol η bind Rev1) can form and that these tri-partite complexes exchanged rapidly (91). This very flexible dynamic may allow for the rapid sampling of TLS polymerases at a damage site to select the activity most appropriate.
When and where does TLS occur? Many models assume the polymerases switch at the stalled fork in order for TLS to occur. Recently another pathway of DNA damage tolerance has been discovered, direct lesion bypass by the replicative polymerase itself in the replisome (Fig. 1a to 1c) (92). Using the reconstituted E. coli replication system and damage-containing templates (46) as described above, it was noted that the Pol III HE (prepared from a polB dinB umuC negative strain and with the proofreading exonuclease fully active) itself could bypass both a CPD and an abasic site analog. Bypass efficiency was proportional to deoxynucleotide concentrations equivalent to those found in vivo and was dependent on the frequency of primer synthesis downstream of the lesion. Translesion synthesis came at the expense of lesion-skipping replication restart. Thus, the stalled leading-strand polymerase was likely cycling between proofreading and polymerization at the 3′-end of the nascent leading strand. Whereas polymerization across from the CPD will be slow compared to the normal chemical step, it was still faster than the amount of time required for progression of the DnaB helicase downstream and the synthesis of a new leading-strand primer to enable the lesion skip. Remarkably, bypass only occurred when the Pol III HE was integrated in the replisome. The enzyme itself could not bypass template damage in a primer-extension assay. These findings suggest that DNA damage at the replication fork can be replicated directly by the replisome without the need to activate error-prone pathways.
How does this observation impact our thinking about TLS? The increase in spontaneous mutation in E. coli cells that have been UV-irradiated is termed either UV- or SOS-mutagenesis. This phenomenon, discovered by Witkin (93), is inextricably linked to TLS. Subsequent early studies suggested that the polymerase subunit (encoded by dnaE) of the Pol III HE was involved in the process. Bridges et al. (94) reported that a function of dnaE was required for UV-induced mutagenesis. The discovery that umuC and umuD were required for UV-mutagenesis (68, 69) led to the hypothesis that these gene products were somehow modifying the Pol III HE to allow TLS, perhaps by inactivating the proofreading subunit ε or directly modifying the α subunit (95, 96). However, the subsequent discovery that UmuC was a TLS DNA polymerase (66, 67), directed attention away from the Pol III HE as the possible agent of mutagenesis. Nevertheless, Moses’ group (97) showed that the dnaE gene product itself was required for UV mutagenesis. How the Pol III HE participated in UV-mutagenesis was therefore left unresolved.
The current study (92) show that the Pol III HE can perform TLS under conditions similar to those found under SOS induction, where it has been reported that nucleotide concentrations increase 2- to 4-fold (98). Furthermore, the frequency of base substitutions increases during Pol III HE-catalyzed TLS at the template position prior to the CPD and in the first position opposite the CPD. These observations suggest that UV mutagenesis may occur in two distinct modes: Mutagenesis that occurs directly at the replication fork is the product of base substitutions generated by the action of Pol III HE TLS, whereas mutagenesis resulting from the action of Pol V occurs in the gaps left behind when the replisome performs lesion skipping. These observations therefore suggest that the bulk of specialized TLS polymerase bypass occurs behind the replication fork in the gaps left behind by lesion skipping. In support of this argument, Robinson et al. (99) did not observe co-localization of Pol V (fluorescently tagged UmuC) with replisome foci (fluorescently tagged τ subunit of the Pol III HE) after UV-irradiation.
Does replisome-mediated bypass of template damage happen in eukaryotic cells? Which DNA polymerase would be most likely to be an agent of TLS? As outlined above, because of a high tendency to dissociate from PCNA, it is unlikely that Pol ε would be able to bypass template damage. Similarly, given the inherent primer synthesis capacity of Pol α/primase, this polymerase is more likely to just release from the template at the block and re-bind ahead of it to synthesize a new primer. This leaves Pol δ, which has weak CPD bypass activity in primer extension reactions that is increased by inactivation of the proofreading exonuclease (77, 100, 101). Interestingly, Hirota et al. (102) demonstrated that chicken DT40 cells deficient in the PolD3 subunit of Pol δ were sensitive to DNA damaging agents and had a reduced ability to maintain replication fork progression after UV-irradiation independent of the function of PolD3 as a subunit of Pol ζ. These authors then demonstrated that these defects could be suppressed by eliminating the proofreading exonuclease activity of Pol δ (103). These studies suggest that TLS at or close to the fork might indeed involve Pol δ. Whether such an activity is replisome associated remains to be determined.
3.4. Template Switching and Fork Reversal
A template switch (TS) is a form of DNA repair that is considered a DNA damage tolerance pathway. There are two types of TS’s that have been reported: a TS behind the fork that initiates in the gap opposite a DNA lesion that has been skipped over (Fig. 1a to 1b to 1e to 1f) and a TS via nascent strand regression at the stalled fork to form a reversed fork (Fig. 1a to 1b to 1g to 1h). In each case, the same basic steps are required for such events. There must be some mechanism for unwinding the nascent strands from the template strands and annealing the nascent strands together. This can be accomplished by a DNA helicase, a DNA translocase capable of branch migration, a strand-pairing DNA recombinase, or, in the case of fork regression, the accumulation of positive superhelical tension. A DNA polymerase is then required to extend the stalled nascent DNA along the switched template strand allowing the stalled leading strand to “advance” past the lesion (Fig. 1f and 1h). These models envision a switch of the nascent strands back to their proper template strands after some time. At this point, in the post-replicative model, because the replication fork that initially encountered the lesion has continued downstream, the paired nascent strands can be resolved some time later by a “dissolution” mechanism. On the other hand, with fork reversal, recovery requires either resetting of the nascent strands back to their original configuration (Fig. 1h to 1i) and subsequent restart (or rescue by another replication fork coming from the opposite direction) or an HR-directed strand invasion event of one of the nascent strands in the reversed fork ahead of the damage on the parental duplex (Fig. 1h to 1j), followed possibly by the equivalent of break-induced replication (BIR) until a replication fork coming from the other direction is encountered. The double Holliday junctions left behind could then be resolved later by either a dissolution- or nuclease-based resolution mechanism. There are many different proteins that appear to be involved in these pathways.
Branzei and colleagues (5, 50, 104–107) have taken the lead in arguing that in yeast, if lesion skipping can occur, a TS behind the fork in the gaps left behind is the preferred pathway of DNA damage tolerance. The recombination proteins Rad51, Rad55, and Rad57 are involved in the forming the initial D loop for the template switch with some processing by ExoI possibly required. Pol δ extends the DNA in the nascent DNA duplex formed by the TS and is likely responsible for any additional synthesis required to close the gap in the nascent DNA. The Sgs1-Top3-Rmi1 complex is the responsible for resolving the double Holliday junction formed. That this form of TS occurs after lesion skipping is strongly supported by the observation that mutations that inactivate the primase activity of Pol α/primase or disrupt coupling of Pol α/primase to the MCMs result in a decrease in TS behind the fork and an increase in fork reversal (50).
Rad51 activity is required for either version of TS, but HR tends to be suppressed during DNA replication. In yeast, inhibition of HR during replication is accomplished in large part via SUMOylation of PCNA on Lys164 (108). Several proteins involved in transactions at replication forks bind to SUMO-PCNA via their SUMO interacting motifs and PIPs. The best studied example is the UvrD-like DNA helicase Srs2 in yeast (109), which is an anti-recombinase by virtue of its ability to disrupt Rad51-ssDNA filaments (110, 111). Recently, it was shown that Srs2 at stalled forks can be degraded via the engagement of Esc2, which contains two SUMO-like domains (112). This finding suggests that a temporal window could be opened at the stalled fork that allowed access of Rad51. Presumably this window is closed by either increased PCNA SUMOylation or its polyubiquitylation on Lys164 that favors TS.
The situation in metazoans is similar, but more involved. Here, a number of different proteins have been reported to suppress HR at stalled forks. SUMO-PCNA plays a role as an attractant for PARI (113, 114), which, like Srs2, contains SUMO interacting motif-, PIP-, and UvrD helicase-like domains, although PARI anti-recombinase activity does not require the latter (114). Two members of the RecQ-family of DNA helicases in human cells, BLM and RECQL5, have recombination suppression activity, presumably by disrupting Rad51-ssDNA filaments (115, 116). RECQL5 interacts with PCNA and is a presumed component of the replication machinery (117). BLM polyubiquitylation on Lys63 as a result of hydroxyurea treatment draws it to the stalled fork via an interaction with RAP80 (118). Suppression of SUMOylation of RPA by the SUMO protease SENP6 also appears to suppress HR, possibly by preventing recruitment of Rad51 (119). A new protein, RADX, with an RPA-like ssDNA-binding activity, was identified and shown to modulate RAD51 activity by binding to ssDNA at stalled forks. It was proposed that this attribute prevents over-accumulation of Rad51 at stalled forks (120).
The second TS, replication fork reversal, was first noted as a potential DNA damage repair pathway by Higgins et al. (121). That fork reversal occurred was first established in E. coli, primarily by Michel and colleagues (6, 122). The agents of regression of the stalled fork could be the DNA helicase RecG, which seems mechanistically to be especially well suited to regressing stalled forks (123), the branch migration protein RuvAB, and the strand exchange protein RecA. Fork reversal could occur under many different circumstances [see ref (6)] and the regressed fork processed in a number of different ways. In the example shown in Fig. 1, the nascent leading strand can be extended using the nascent lagging strand as a template (Fig. 1g to 1h). This species could then be reset by reversal of regression to a stalled fork where the nascent strands had both moved past the damage (Fig. 1i). Alternatively, processing of the nascent DNA duplex double-stranded end in the reversed fork could generate a 3′-ended single strand that could invade the parental duplex downstream of the damage (Fig. 1h to 1j). In a different branch of the pathway, once regression has occurred, the damage can be removed by nucleotide excision repair, the Holliday junction in the regressed fork cleaved by a resolvase, and double strand end-processing and strand invasion take place at a position that is upstream of the initial site of template damage (Fig. 1g to 1k to 1l to 1m). Interestingly, most established cases of fork regression in E. coli are not triggered by DNA template damage—use the template damage “triangle” in Fig. 1 as the reference point of fork stalling; the potential pathways are identical—but are caused by replisome collapse, nucleotide deprivation, head-on collisions with transcription complexes, or collisions with blocking proteins (6, 122).
Contemporaneously there was little evidence that fork reversal occurred in eukaryotes. In yeast, fork reversal was shown to occur when the S-phase checkpoint was defective (29, 124). PARP-dependent fork reversal was then demonstrated in both yeast and mammalian cells as a result of treatment with camptothecin, a topoisomerase I inhibitor that traps the topoisomerase-DNA covalent complex (125). Subsequently, Zellweger et al. (126) observed RAD51-dependent fork reversal in human cancer cell lines upon treatment with a wide spectrum of DNA-damaging and fork stalling agents. These observations, scored by EM examination of psoralen-crosslinked replication intermediates, suggested, remarkably, that 500–4000 reversed forks could exist per cell when treated thusly. Only some of the treatments used induced the S phase checkpoint. The authors concluded that fork reversal was a global response to cellular replication stress. It should be noted, though, that the cell line used for most of these studies, U20S, has altered expression of both p16-INK4A and p14-ARF (127), inhibitors of the G1-S transition, as well as an altered DNA damage response (128).
Whereas RAD51 was shown to be the agent of fork reversal in the studies described above from Lopes and colleagues, there are many other potential candidates in mammalian cells [see (7) for a recent review of the topic], including FBH1; the RecQ-like DNA helicases BLM, WRN, and RECQ5; the SWI/SNF protein family members RAD5, RAD54, SMARCAL1, and ZRANB3; and FANCM, all of which may act independently or in cooperation with RAD51.
Nascent DNA at stalled forks in mammalian cells appears to be targeted for degradation by the MRE11 nuclease in the MRN complex (129–134). BRCA2-dependent, RAD51-ssDNA filaments prevents this DNA degradation and a reversed fork often appears in schematics of these fork stabilization pathways (135). The involvement of metazoan recombination proteins in protecting nascent DNA from degradation was foreshadowed by similar studies in E. coli demonstrating that RecA, RecF, RecO, and RecR protected nascent DNA at stalled forks from digestion by a combination of the RecJ nuclease and the RecQ DNA helicase (136)
Fork reversal requires that the 3′-ends of both the nascent leading and lagging strands must be available to freely rotate about the template strands. Thus, at the very least, it would seem that the respective DNA polymerases would have to release from the primer-templates. In studies using the reconstituted E. coli replication system described above with a CPD in the leading-strand template, fork reversal by RecG and RuvAB could be observed even when the replisome proteins were still associated with the template (137). It seems unlikely that either the helicase DnaB or the lagging-strand polymerase would be an impediment to fork reversal. These enzymes continue to migrate downstream away from the stalled leading-strand polymerase (33). It was suggested that fork reversal ocurred after the leading-strand polymerase had cycled forward to a new primer on the leading-strand template in the lesion-skipping reaction and thus was, from this point of view, a post-replicative event. Template regression can be initated at a gap in a nascent strand.
A similar scenario could present in eukaryotic cells with the potential lesion-skipping reaction at template damage as described above. Fork blocking proteins, drug treatments that reduce the deoxynucleotide pool or inhibit the DNA polymerases, and accumulated positive superhelical tension presents a different situation, however. These impediments likely inhibit any fork progression. So how does fork reversal occur? One simple solution is collapse of the replisome and dissociation of its components, or at least the polymerases. Studies using the iPOND technique (138) of protein composition at stalled forks in mammalian cells indicate, however, that many replisome components appear to be preserved (139, 140). Thus it is possible that the remodelers of strands at stalled forks may also act themselves, or in concert with other proteins, to displace replisome components from the stalled fork region, yet still allow them to remain on the DNA. Indeed, single molecule studies of fork stalling and reversal using Bacteriophage T4 proteins demonstrated that the UvsW recombination protein could regress the fork while displacing the DNA polymerase holoenzyme to the nascent DNA regressed arm (141).
4. Replication Restart
In the absence of lesion skipping, fork remodeling creates a potential demand for origin-independent replication restart. Such restart can be driven by HR (Fig. 1j and 1m) in which case whereas DNA polymerases may have direct access to the 3′-ends of the nascent DNA to continue synthesis, there is still a demand for re-loading the DNA helicase to form a replication fork. Or restart can occur at a fork whose proper structure has been restored (Fig. 1j). How these reactions occur in bacterial cells is clear, these pathways are less clear in eukaryotes.
Both HR-directed and origin-independent replication restart in E. coli is executed by the restart primosomal proteins PriA, PriB, PriC, and DnaT [reviewed recently in (6)]. These proteins can load the DnaB helicase to either a D-loop (142) or, in a manner dependent on the disposition of the nascent strands, directly to a reset stalled fork (143). Once DnaB is on the DNA it can bind the primase, DnaG, that will synthesize a primer for a new Okazaki fragment. The 3′-end of the nascent leading strand can be used as the primer for resumption of leading-strand synthesis. The presence of these free 3′-ends draws the DNA Pol III HE to the DNA to re-establish the replisome. It is also possible that DnaG itself re-establishes leading-strand synthesis by forming a primer on the leading-strand template as it does during the lesion skipping reaction (46, 144).
With many more origins licensed than active in mammalian cells, the necessity of replication restart is reduced significantly compared to the imperative for it in bacteria. Indeed, dormant origins are required for survival of human cancer cells during replication stress (145, 146) and to suppress fork stalling during normal S phase (147). Nevertheless, many studies of DNA fiber analyses indicate that restart does occur in mammalian cells. As discussed above, some fraction of the observed restart is either Prim/Pol-mediated (lesion skipping and catch up) or HR-mediated (Fig. 1j and 1m), dependent on RAD51-catalyzed strand invasion either ahead of the template damage or with the completely replicated sister duplex. Replisome re-assembly under these conditions may reflect a BIR-type of mechanism. Whereas it is known that a number of replisome components participate in BIR, how these components are assembled into a functional “BIR-replisome” is unclear. And both Mcm2–7 and Pif1 have been argued to be the DNA helicases present at the migrating D loop (148). Presumably, if replication from these recombination intermediates is of the BIR mode, it does not go on for long. Such replication is likely to be mutagenic and either rescued by a bona fide replication fork coming from the opposite direction or terminated by cleavage of the recombination intermediate by Mus81 (59). Interestingly, studies in Xenopus cell extracts showed that after replication fork collapse at a single-strand break, the CMG complex loses GINS and Pol ε, which are then re-loaded in an origin-independent manner dependent on MRE11 and RAD51 (149). However, the actual mechanism is unknown.
Regressed forks can be reset to the standard configuration in mammalian cells by exonucleolytic digestion by a combination of WRN and DNA2 (150) and reversal of the regressed fork by either RECQ1 (126, 151) or SMARCAL1 (152). It is unclear whether these reset forks can restart replication. There is no known ORC-independent helicase loading mechanism in eukaryotes. Further studies will be required to answer this question.
5. CONCLUSIONS
Lesions in the DNA template are not the absolute impediment to replication fork progression that they were initially thought to be. Replisomes can jump over them, replicate across them, and make up the lost ground by switching template strands. A new dynamic view of how replisomes operate may reflect evolutionarily built-in tolerance mechanisms for template damage. By contrast, obstructions that stop replication fork progression completely require more elaborate mechanisms to sustain complete replication of the genome. Though not covered in this review, transcription-replication conflicts seem to be more destabilizing to the genome than single base alterations. In this vein, it would be remiss not to cite two very recent important reports demonstrating the central role of RNA-DNA hybrid formation in destabilizing the genome during head-on replication-transcription collisions (153, 154).
ACKNOWLEDGMENTS
Thanks to Xiaolan Zhao and John Petrini for a critical reading of the manuscript and to John for drawing figure 1. Studies from the author’s laboratory were supported by NIH grant GM34557 and NCI Comprehensive Cancer Center Support Grant P30CA008748 to Craig Thompson.
Footnotes
DISCLOSURE STATEMENT
The author declares no financial interests or conflicts of interest.
LITERATURE CITED
- 1.Aguilera A, Garcia-Muse T. 2013. Causes of genome instability. Annu. Rev. Genet 47:1–32 [DOI] [PubMed] [Google Scholar]
- 2.Cha HJ, Yim H. 2013. The accumulation of DNA repair defects is the molecular origin of carcinogenesis. Tumour Biol. 34:3293–302 [DOI] [PubMed] [Google Scholar]
- 3.Macheret M, Halazonetis TD. 2015. DNA replication stress as a hallmark of cancer. Ann Re Patho 10:425–48 [DOI] [PubMed] [Google Scholar]
- 4.Yeeles JT, Poli J, Marians KJ, Pasero P. 2013. Rescuing stalled or damaged replication forks. Cold Spring Harb. Perspect. Biol 5:271–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Branzei D, Szakal B. 2017. Building up and breaking down: mechanisms controlling recombination during replication. Crit. Rev. Biochem. Mol. Biol 52:381–94 [DOI] [PubMed] [Google Scholar]
- 6.Michel B, Sandler SJ. 2017. Replication restart in bacteria. J. Bacteriol 199:e00102–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neelsen KJ, Lopes M. 2015. Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat. Rev. Mol. Cell Biol 16:207–20 [DOI] [PubMed] [Google Scholar]
- 8.Hedglin M, Benkovic SJ. 2017. Eukaryotic translesion DNA synthesis on the leading and lagging strands: Unique detours around the same obstacle. Chem. Rev 117:7857–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Techer H, Koundrioukoff S, Nicolas A, Debatisse M. 2017. The impact of replication stress on replication dynamics and DNA damage in vertebrate cells. Nat. Rev. Genet 18:535–50 [DOI] [PubMed] [Google Scholar]
- 10.Yazinski SA, Zou L. 2016. Functions, Regulation, and therapeutic implications of the ATR checkpoint pathway. Annu. Rev. Genet 50:155–73 [DOI] [PubMed] [Google Scholar]
- 11.Toledo L, Neelsen KJ, Lukas J. 2017. Replication catastrophe: When a checkpoint fails because of exhaustion. Mol. Cell 66:735–49 [DOI] [PubMed] [Google Scholar]
- 12.Okazaki R, Okazaki T, Sakabe K, Sugimoto K, Sugino A. 1968. Mechanism of DNA chain growth. I. Possible discontinuity and unusual secondary structure of newly synthesized chains. PNAS 59:598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McHenry CS. 2011. DNA replicases from a bacterial perspective. Annu. Rev. Biochem 80:403–36 [DOI] [PubMed] [Google Scholar]
- 14.Bleichert F, Botchan MR, Berger JM. 2017. Mechanisms for initiating cellular DNA replication. Science 355: [DOI] [PubMed] [Google Scholar]
- 15.Georgescu RE, Schauer GD, Yao NY, Langston LD, Yurieva O, et al. 2015. Reconstitution of a eukaryotic replisome reveals suppression mechanisms that define leading/lagging strand operation. Elife 4:e04988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yeeles JT, Deegan TD, Janska A, Early A, Diffley JF. 2015. Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 519:431–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yeeles JT, Janska A, Early A, Diffley JF. 2017. How the eukaryotic replisome achieves rapid and efficient DNA replication. Mol. Cell 65:105–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurat CF, Yeeles JT, Patel H, Early A, Diffley JF. 2017. Chromatin controls DNA replication origin selection, lagging-strand synthesis, and replication fork rates. Mol. Cell 65:117–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Devbhandari S, Jiang J, Kumar C, Whitehouse I, Remus D. 2017. Chromatin constrains the initiation and elongation of DNA replication. Mol. Cell 65:131–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burgers PMJ, Kunkel TA. 2017. Eukaryotic DNA replication fork. Annu. Rev. Biochem 86:417–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Georgescu R, Yuan Z, Bai L, de Luna Almeida Santos R, Sun J, et al. 2017. Structure of eukaryotic CMG helicase at a replication fork and implications to replisome architecture and origin initiation. PNAS 114:E697–E706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang L, Davey MJ, O’Donnell M. 1999. Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol. Cell 4:541–53 [DOI] [PubMed] [Google Scholar]
- 23.Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, et al. 2006. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol 8:358–66 [DOI] [PubMed] [Google Scholar]
- 24.McInerney P, O’Donnell M. 2004. Functional uncoupling of twin polymerases: mechanism of polymerase dissociation from a lagging-strand block. J. Biol. Chem 279:21543–51 [DOI] [PubMed] [Google Scholar]
- 25.Higuchi K, Katayama T, Iwai S, Hidaka M, Horiuchi T, Maki H. 2003. Fate of DNA replication fork encountering a single DNA lesion during oriC plasmid DNA replication in vitro. Genes Cells 8:437–49 [DOI] [PubMed] [Google Scholar]
- 26.Pages V, Fuchs RP. 2003. Uncoupling of leading- and lagging-strand DNA replication during lesion bypass in vivo. Science 300:1300–3 [DOI] [PubMed] [Google Scholar]
- 27.Lopes M, Foiani M, Sogo JM. 2006. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell 21:15–27 [DOI] [PubMed] [Google Scholar]
- 28.Sassanfar M, Roberts JW. 1990. Nature of the SOS-inducing signal in Escherichia coli. The involvement of DNA replication. J. Mol. Biol 212:79–96 [DOI] [PubMed] [Google Scholar]
- 29.Sogo JM, Lopes M, Foiani M. 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297:599–602 [DOI] [PubMed] [Google Scholar]
- 30.Cha RS, Kleckner N. 2002. ATR homolog Mec1 promotes fork progression, thus averting breaks in replication slow zones. Science 297:602–6 [DOI] [PubMed] [Google Scholar]
- 31.Zou L, Elledge SJ. 2003. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300:1542–8 [DOI] [PubMed] [Google Scholar]
- 32.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. 2005. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19:1040–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeeles JT, Marians KJ. 2013. Dynamics of leading-strand lesion skipping by the replisome. Mol. Cell 52:855–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graham JE, Marians KJ, Kowalczykowski SC. 2017. Independent and stochastic action of DNA polymerases in the replisome. Cell 169:1201–13 e17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pandey M, Syed S, Donmez I, Patel G, Ha T, Patel SS. 2009. Coordinating DNA replication by means of priming loop and differential synthesis rate. Nature 462:940–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee JB, Hite RK, Hamdan SM, Xie XS, Richardson CC, van Oijen AM. 2006. DNA primase acts as a molecular brake in DNA replication. Nature 439:621–4 [DOI] [PubMed] [Google Scholar]
- 37.Langston LD, Zhang D, Yurieva O, Georgescu RE, Finkelstein J, et al. 2014. CMG helicase and DNA polymerase epsilon form a functional 15-subunit holoenzyme for eukaryotic leading-strand DNA replication. PNAS 111:15390–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun J, Shi Y, Georgescu RE, Yuan Z, Chait BT, et al. 2015. The architecture of a eukaryotic replisome. Nat. Struct. Mol. Biol 22:976–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou JC, Janska A, Goswami P, Renault L, Abid Ali F, et al. 2017. CMG-Pol epsilon dynamics suggests a mechanism for the establishment of leading-strand synthesis in the eukaryotic replisome. PNAS 114:4141–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon AC, Zhou JC, Perera RL, van Deursen F, Evrin C, et al. 2014. A Ctf4 trimer couples the CMG helicase to DNA polymerase alpha in the eukaryotic replisome. Nature 510:293–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Villa F, Simon AC, Ortiz Bazan MA, Kilkenny ML, Wirthensohn D, et al. 2016. Ctf4 Is a hub in the eukaryotic replisome that links multiple CIP-box proteins to the CMG helicase. Mol. Cell 63:385–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schauer GD, O’Donnell ME. 2017. Quality control mechanisms exclude incorrect polymerases from the eukaryotic replication fork. PNAS 114:675–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prakash L 1981. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol. Gen. Genet 184:471–8 [DOI] [PubMed] [Google Scholar]
- 44.Lehmann AR. 1972. Post-replication repair of DNA in ultraviolet-irradiated mammalian cells. No gaps in DNA synthesized late after ultraviolet irradiation. Eur. J. Biochem 31:438–45 [DOI] [PubMed] [Google Scholar]
- 45.Rupp WD, Howard-Flanders P. 1968. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J. Mol. Biol 31:291–304 [DOI] [PubMed] [Google Scholar]
- 46.Yeeles JTP, Marians KJ. 2011. The Escherichia coli replisome is inherently DNA damage tolerant. Science 334:235–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Callegari AJ, Clark E, Pneuman A, Kelly TJ. 2010. Postreplication gaps at UV lesions are signals for checkpoint activation. PNAS 107:8219–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Callegari AJ, Kelly TJ. 2006. UV irradiation induces a postreplication DNA damage checkpoint. PNAS 103:15877–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elvers I, Johansson F, Groth P, Erixon K, Helleday T. 2011. UV stalled replication forks restart by re-priming in human fibroblasts. Nucleic Acids Res. 39:7049–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fumasoni M, Zwicky K, Vanoli F, Lopes M, Branzei D. 2015. Error-free DNA damage tolerance and sister chromatid proximity during DNA replication rely on the Polalpha/Primase/Ctf4 Complex. Mol. Cell 57:812–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iyer LM, Koonin EV, Leipe DD, Aravind L. 2005. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 33:3875–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bianchi J, Rudd SG, Jozwiakowski SK, Bailey LJ, Soura V, et al. 2013. PrimPol bypasses UV photoproducts during eukaryotic chromosomal DNA replication. Mol. Cell 52:566–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rudd SG, Glover L, Jozwiakowski SK, Horn D, Doherty AJ. 2013. PPL2 translesion polymerase is essential for the completion of chromosomal DNA replication in the African trypanosome. Mol. Cell 52:554–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wan L, Lou J, Xia Y, Su B, Liu T, et al. 2013. hPrimpol1/CCDC111 is a human DNA primase-polymerase required for the maintenance of genome integrity. EMBO Rep. 14:1104–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Garcia-Gomez S, Reyes A, Martinez-Jimenez MI, Chocron ES, Mouron S, et al. 2013. PrimPol, an archaic primase/polymerase operating in human cells. Mol. Cell 52:541–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mouron S, Rodriguez-Acebes S, Martinez-Jimenez MI, Garcia-Gomez S, Chocron S, et al. 2013. Repriming of DNA synthesis at stalled replication forks by human PrimPol. Nat. Struct. Mol. Biol 20:1383–9 [DOI] [PubMed] [Google Scholar]
- 57.Kobayashi K, Guilliam TA, Tsuda M, Yamamoto J, Bailey LJ, et al. 2016. Repriming by PrimPol is critical for DNA replication restart downstream of lesions and chain-terminating nucleosides. Cell Cycle 15:1997–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lydeard JR, Jain S, Yamaguchi M, Haber JE. 2007. Break-induced replication and telomerase-independent telomere maintenance require Pol32. Nature 448:820–3 [DOI] [PubMed] [Google Scholar]
- 59.Mayle R, Campbell IM, Beck CR, Yu Y, Wilson M, et al. 2015. DNA REPAIR. Mus81 and converging forks limit the mutagenicity of replication fork breakage. Science 349:742–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miyabe I, Mizuno K, Keszthelyi A, Daigaku Y, Skouteri M, et al. 2015. Polymerase delta replicates both strands after homologous recombination-dependent fork restart. Nat. Struct. Mol. Biol 22:932–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, et al. 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424:1078–83 [DOI] [PubMed] [Google Scholar]
- 62.Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K. 2005. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 19:1905–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang W, Woodgate R. 2007. What a difference a decade makes: insights into translesion DNA synthesis. PNAS 104:15591–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McCulloch SD, Kunkel TA. 2008. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 18:148–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wagner J, Gruz P, Kim SR, Yamada M, Matsui K, et al. 1999. The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol. Cell 4:281–86 [DOI] [PubMed] [Google Scholar]
- 66.Tang M, Shen X, Frank EG, O’Donnell M, Woodgate R, Goodman MF. 1999. UmuD’(2)C is an error-prone DNA polymerase, Escherichia coli pol V. PNAS 96:8919–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Reuven NB, Arad G, Maor-Shoshani A, Livneh Z. 1999. The mutagenesis protein UmuC is a DNA polymerase activated by UmuD’, RecA, and SSB and is specialized for translesion replication. J. Biol. Chem 274:31763–6 [DOI] [PubMed] [Google Scholar]
- 68.Kato T, Shinoura Y. 1977. Isolation and characterization of mutants of Escherichia coli deficient in induction of mutations by ultraviolet light. Mol. Gen. Genet 156:121–31 [DOI] [PubMed] [Google Scholar]
- 69.Steinborn G 1978. Uvm mutants of Escherichia coli K12 deficient in UV mutagenesis. I. Isolation of uvm mutants and their phenotypical characterization in DNA repair and mutagenesis. Mol. Gen. Genet 165:87–93 [DOI] [PubMed] [Google Scholar]
- 70.Goodman MF, Woodgate R. 2013. Translesion DNA polymerases. Cold Spring Harb. Perspect. Biol 5:a010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Livneh Z, Ziv O, Shachar S. 2010. Multiple two-polymerase mechanisms in mammalian translesion DNA synthesis. Cell Cycle 9:729–35 [DOI] [PubMed] [Google Scholar]
- 72.Gao Y, Mutter-Rottmayer E, Zlatanou A, Vaziri C, Yang Y. 2017. Mechanisms of post-replication DNA repair. Genes 8:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao L, Washington MT. 2017. Translesion synthesis: Insights into the selection and switching of DNA Polymerases. Genes (Basel) 8:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Indiani C, McInerney P, Georgescu R, Goodman MF, O’Donnell M. 2005. A sliding-clamp toolbelt binds high- and low-fidelity DNA polymerases simultaneously. Mol. Cell 19:805–15 [DOI] [PubMed] [Google Scholar]
- 75.Indiani C, Langston LD, Yurieva O, Goodman MF, O’Donnell M. 2009. Translesion DNA polymerases remodel the replisome and alter the speed of the replicative helicase. PNAS 106:6031–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kath JE, Jergic S, Heltzel JM, Jacob DT, Dixon NE, et al. 2014. Polymerase exchange on single DNA molecules reveals processivity clamp control of translesion synthesis. PNAS 111:7647–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McCulloch SD, Kokoska RJ, Chilkova O, Welch CM, Johansson E, et al. 2004. Enzymatic switching for efficient and accurate translesion DNA replication. Nucleic Acids Res. 32:4665–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fernandez-Leiro R, Conrad J, Scheres SH, Lamers MH. 2015. cryo-EM structures of the E. coli replicative DNA polymerase reveal its dynamic interactions with the DNA sliding clamp, exonuclease and tau. Elife 4:e11134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lewis JS, Spenkelink LM, Jergic S, Wood EA, Monachino E, et al. 2017. Single-molecule visualization of fast polymerase turnover in the bacterial replisome. Elife 6:e23932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yuan Q, Dohrmann PR, Sutton MD, McHenry CS. 2016. DNA polymerase III, but not polymerase IV, must be bound to a tau-containing DnaX complex to enable exchange into replication forks. J. Biol. Chem 291:11727–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Beattie TR, Kapadia N, Nicolas E, Uphoff S, Wollman AJ, et al. 2017. Frequent exchange of the DNA polymerase during bacterial chromosome replication. Elife 6:e21763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sale JE. 2012. Competition, collaboration and coordination--determining how cells bypass DNA damage. J. Cell Sci 125:1633–43 [DOI] [PubMed] [Google Scholar]
- 83.Xu X, Blackwell S, Lin A, Li F, Qin Z, Xiao W. 2015. Error-free DNA-damage tolerance in Saccharomyces cerevisiae. Mutat. Res. Rev 764:43–50 [DOI] [PubMed] [Google Scholar]
- 84.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419:135–41 [DOI] [PubMed] [Google Scholar]
- 85.Bienko M, Green CM, Crosetto N, Rudolf F, Zapart G, et al. 2005. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310:1821–24 [DOI] [PubMed] [Google Scholar]
- 86.Hedglin M, Pandey B, Benkovic SJ. 2016. Characterization of human translesion DNA synthesis across a UV-induced DNA lesion. Elife 5:e19788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guo C, Sonoda E, Tang TS, Parker JL, Bielen AB, et al. 2006. REV1 protein interacts with PCNA: significance of the REV1 BRCT domain in vitro and in vivo. Mol. Cell 23:265–71 [DOI] [PubMed] [Google Scholar]
- 88.Pustovalova Y, Maciejewski MW, Korzhnev DM. 2013. NMR mapping of PCNA interaction with translesion synthesis DNA polymerase Rev1 mediated by Rev1-BRCT domain. J. Mol. Biol 425:3091–105 [DOI] [PubMed] [Google Scholar]
- 89.Sharma NM, Kochenova OV, Shcherbakova PV. 2011. The non-canonical protein binding site at the monomer-monomer interface of yeast proliferating cell nuclear antigen (PCNA) regulates the Rev1-PCNA interaction and Polzeta/Rev1-dependent translesion DNA synthesis. J. Biol. Chem 286:33557–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boehm EM, Powers KT, Kondratick CM, Spies M, Houtman JC, Washington MT. 2016. The Proliferating Cell Nuclear Antigen (PCNA)-interacting protein (PIP) motif of DNA polymerase eta mediates its interaction with the C-terminal domain of Rev1. J. Biol. Chem 291:8735–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Boehm EM, Spies M, Washington MT. 2016. PCNA tool belts and polymerase bridges form during translesion synthesis. Nucleic Acids Res. 44:8250–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Nevin P, Gabbai CC, Marians KJ. 2017. Replisome-mediated translesion synthesis by a cellular replicase. J. Biol. Chem 292:13883–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Witkin EM. 1976. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol. Rev 40:869–907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bridges BA, Mottershead RP. 1976. Mutagenic DNA repair in Escherichia coli. III. Requirement for a function of DNA polymerase III in ultraviolet-light mutagenesis. Mol. Gen. Genet 144:53–8 [DOI] [PubMed] [Google Scholar]
- 95.Lu C, Scheuermann RH, Echols H. 1986. Capacity of RecA protein to bind preferentially to UV lesions and inhibit the editing subunit (epsilon) of DNA polymerase III: a possible mechanism for SOS-induced targeted mutagenesis. PNAS 83:619–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Rajagopalan M, Lu C, Woodgate R, O’Donnell M, Goodman MF, Echols H. 1992. Activity of the purified mutagenesis proteins UmuC, UmuD’, and RecA in replicative bypass of an abasic DNA lesion by DNA polymerase III. PNAS 89:10777–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hagensee ME, Timme TL, Bryan SK, Moses RE. 1987. DNA polymerase III of Escherichia coli is required for UV and ethyl methanesulfonate mutagenesis. PNAS 84:4195–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gon S, Napolitano R, Rocha W, Coulon S, Fuchs RP. 2011. Increase in dNTP pool size during the DNA damage response plays a key role in spontaneous and induced-mutagenesis in Escherichia coli. PNAS 108:19311–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Robinson A, McDonald JP, Caldas VE, Patel M, Wood EA, et al. 2015. Regulation of mutagenic DNA polymerase V activation in space and time. PLoS Genet. 11:e1005482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Meng X, Zhou Y, Zhang S, Lee EY, Frick DN, Lee MY. 2009. DNA damage alters DNA polymerase delta to a form that exhibits increased discrimination against modified template bases and mismatched primers. Nucleic Acids Res. 37:647–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Narita T, Tsurimoto T, Yamamoto J, Nishihara K, Ogawa K, et al. 2010. Human replicative DNA polymerase delta can bypass T-T (6–4) ultraviolet photoproducts on template strands. Genes Cells 15:1228–39 [DOI] [PubMed] [Google Scholar]
- 102.Hirota K, Yoshikiyo K, Guilbaud G, Tsurimoto T, Murai J, et al. 2015. The POLD3 subunit of DNA polymerase delta can promote translesion synthesis independently of DNA polymerase zeta. Nucleic Acids Res. 43:1671–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hirota K, Tsuda M, Mohiuddin, Tsurimoto T, Cohen IS, et al. 2016. In vivo evidence for translesion synthesis by the replicative DNA polymerase delta. Nucleic Acids Res. 44:7242–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Vanoli F, Fumasoni M, Szakal B, Maloisel L, Branzei D. 2010. Replication and recombination factors contributing to recombination-dependent bypass of DNA lesions by template switch. PLoS Genet. 6:e1001205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bonner JN, Choi K, Xue X, Torres NP, Szakal B, et al. 2016. Smc5/6 mediated sumoylation of the Sgs1-Top3-Rmi1 complex promotes removal of recombination intermediates. Cell Rep. 16:368–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Branzei D, Szakal B. 2016. DNA damage tolerance by recombination: Molecular pathways and DNA structures. DNA Repair 44:68–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Giannattasio M, Zwicky K, Follonier C, Foiani M, Lopes M, Branzei D. 2014. Visualization of recombination-mediated damage bypass by template switching. Nat. Struct. Mol. Biol 21:884–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pfander B, Moldovan GL, Sacher M, Hoege C, Jentsch S. 2005. SUMO-modified PCNA recruits Srs2 to prevent recombination during S phase. Nature 436:428–33 [DOI] [PubMed] [Google Scholar]
- 109.Armstrong AA, Mohideen F, Lima CD. 2012. Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature 483:59–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, et al. 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423:305–9 [DOI] [PubMed] [Google Scholar]
- 111.Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423:309–12 [DOI] [PubMed] [Google Scholar]
- 112.Urulangodi M, Sebesta M, Menolfi D, Szakal B, Sollier J, et al. 2015. Local regulation of the Srs2 helicase by the SUMO-like domain protein Esc2 promotes recombination at sites of stalled replication. Genes Dev. 29:2067–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Moldovan GL, Dejsuphong D, Petalcorin MI, Hofmann K, Takeda S, et al. 2012. Inhibition of homologous recombination by the PCNA-interacting protein PARI. Mol. Cell 45:75–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Burkovics P, Dome L, Juhasz S, Altmannova V, Sebesta M, et al. 2016. The PCNA-associated protein PARI negatively regulates homologous recombination via the inhibition of DNA repair synthesis. Nucleic Acids Res. 44:3176–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, et al. 2007. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 21:3073–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bugreev DV, Yu X, Egelman EH, Mazin AV. 2007. Novel pro- and anti-recombination activities of the Bloom’s syndrome helicase. Genes Dev. 21:3085–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kanagaraj R, Saydam N, Garcia PL, Zheng L, Janscak P. 2006. Human RECQ5beta helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 34:5217–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tikoo S, Madhavan V, Hussain M, Miller ES, Arora P, et al. 2013. Ubiquitin-dependent recruitment of the Bloom syndrome helicase upon replication stress is required to suppress homologous recombination. EMBO J. 32:1778–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dou H, Huang C, Singh M, Carpenter PB, Yeh ET. 2010. Regulation of DNA repair through deSUMOylation and SUMOylation of replication protein A complex. Mol. Cell 39:333–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dungrawala H, Bhat KP, Le Meur R, Chazin WJ, Ding X, et al. 2017. RADX promotes genome stability and modulates chemosensitivity by regulating RAD51 at replication forks. Mol. Cell 67:374–86 e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Higgins NP, Kato K, Strauss B. 1976. A model for replication repair in mammalian cells. J. Mol. Biol 101:417–25 [DOI] [PubMed] [Google Scholar]
- 122.Atkinson J, McGlynn P. 2009. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 37:3475–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Singleton MR, Scaife S, Wigley DB. 2001. Structural analysis of DNA replication fork reversal by RecG. Cell 107:79–89 [DOI] [PubMed] [Google Scholar]
- 124.Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, et al. 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412:557–61 [DOI] [PubMed] [Google Scholar]
- 125.Ray Chaudhuri A, Hashimoto Y, Herrador R, Neelsen KJ, Fachinetti D, et al. 2012. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat. Struct. Mol. Biol 19:417–23 [DOI] [PubMed] [Google Scholar]
- 126.Zellweger R, Dalcher D, Mutreja K, Berti M, Schmid JA, et al. 2015. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol 208:563–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Park YB, Park MJ, Kimura K, Shimizu K, Lee SH, Yokota J. 2002. Alterations in the INK4a/ARF locus and their effects on the growth of human osteosarcoma cell lines. Cancer Genet. Cytogenet 133:105–11 [DOI] [PubMed] [Google Scholar]
- 128.Lovejoy CA, Li W, Reisenweber S, Thongthip S, Bruno J, et al. 2012. Loss of ATRX, genome instability, and an altered DNA damage response are hallmarks of the alternative lengthening of telomeres pathway. PLoS Genet. 8:e1002772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hashimoto Y, Ray Chaudhuri A, Lopes M, Costanzo V. 2010. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol 17:1305–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. 2011. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145:529–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Schlacher K, Wu H, Jasin M. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 22:106–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ying S, Hamdy FC, Helleday T. 2012. Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res. 72:2814–21 [DOI] [PubMed] [Google Scholar]
- 133.Vallerga MB, Mansilla SF, Federico MB, Bertolin AP, Gottifredi V. 2015. Rad51 recombinase prevents Mre11 nuclease-dependent degradation and excessive PrimPol-mediated elongation of nascent DNA after UV irradiation. PNAS 112:E6624–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kolinjivadi AM, Sannino V, De Antoni A, Zadorozhny K, Kilkenny M, et al. 2017. Smarcal1-mediated fork reversal triggers Mre11-dependent degradation of nascent DNA in the absence of Brca2 and stable Rad51 nucleofilaments. Mol. Cell 67:867–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kolinjivadi AM, Sannino V, de Antoni A, Techer H, Baldi G, Costanzo V. 2017. Moonlighting at replication forks - a new life for homologous recombination proteins BRCA1, BRCA2 and RAD51. FEBS Lett. 591:1083–100 [DOI] [PubMed] [Google Scholar]
- 136.Courcelle J, Donaldson JR, Chow KH, Courcelle CT. 2003. DNA damage-induced replication fork regression and processing in Escherichia coli. Science 299:1064–7 [DOI] [PubMed] [Google Scholar]
- 137.Gupta S, Yeeles JTP, and Marians KJ 2014. Regression of replication forks stalled by leading-strand template damage I. Both RecG and RuvAB catalyze regression, but RuvC cleaves the Holliday junctions formed by RecG preferentially. J. Biol. Chem 289:28376–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sirbu BM, Couch FB, Feigerle JT, Bhaskara S, Hiebert SW, Cortez D. 2011. Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 25:1320–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lopez-Contreras AJ, Ruppen I, Nieto-Soler M, Murga M, Rodriguez-Acebes S, et al. 2013. A proteomic characterization of factors enriched at nascent DNA molecules. Cell Rep. 3:1105–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lossaint G, Larroque M, Ribeyre C, Bec N, Larroque C, et al. 2013. FANCD2 binds MCM proteins and controls replisome function upon activation of s phase checkpoint signaling. Mol. Cell 51:678–90 [DOI] [PubMed] [Google Scholar]
- 141.Manosas M, Perumal SK, Croquette V, Benkovic SJ. 2012. Direct observation of stalled fork restart via fork regression in the T4 replication system. Science 338:1217–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Xu L, Marians KJ. 2003. PriA mediates DNA replication pathway choice at recombination intermediates. Mol. Cell 11:817–26 [DOI] [PubMed] [Google Scholar]
- 143.Heller RC, Marians KJ. 2005. The disposition of nascent strands at stalled replication forks dictates the pathway of replisome loading during restart. Mol. Cell 17:733–43 [DOI] [PubMed] [Google Scholar]
- 144.Heller RC, Marians KJ. 2006. Replication fork reactivation downstream of a blocked nascent leading strand. Nature 439:557–62 [DOI] [PubMed] [Google Scholar]
- 145.Ibarra A, Schwob E, Mendez J. 2008. Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. PNAS 105:8956–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Ge XQ, Jackson DA, Blow JJ. 2007. Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes Dev. 21:3331–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kawabata T, Luebben SW, Yamaguchi S, Ilves I, Matise I, et al. 2011. Stalled fork rescue via dormant replication origins in unchallenged S phase promotes proper chromosome segregation and tumor suppression. Mol. Cell 41:543–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sakofsky CJ, Malkova A. 2017. Break induced replication in eukaryotes: mechanisms, functions, and consequences. Crit. Rev. Biochem. Mol. Biol 52:395–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hashimoto Y, Puddu F, Costanzo V. 2011. RAD51- and MRE11-dependent reassembly of uncoupled CMG helicase complex at collapsed replication forks. Nat. Struct. Mol. Biol 19:17–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Thangavel S, Berti M, Levikova M, Pinto C, Gomathinayagam S, et al. 2015. DNA2 drives processing and restart of reversed replication forks in human cells. J. Cell Biol 208:545–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Berti M, Ray Chaudhuri A, Thangavel S, Gomathinayagam S, Kenig S, et al. 2013. Human RECQ1 promotes restart of replication forks reversed by DNA topoisomerase I inhibition. Nat. Struct. Mol. Biol 20:347–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Betous R, Couch FB, Mason AC, Eichman BF, Manosas M, Cortez D. 2013. Substrate-selective repair and restart of replication forks by DNA translocases. Cell Rep. 3:1958–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hamperl S, Bocek MJ, Saldivar JC, Swigut T, Cimprich KA. 2017. Transcription-replication conflict orientation modulates R-loop levels and activates distinct DNA damage responses. Cell 170:774–86 e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lang KS, Hall AN, Merrikh CN, Ragheb M, Tabakh H, et al. 2017. Replication-transcription conflicts generate R-loops that orchestrate bacterial stress survival and pathogenesis. Cell 170:787–99 e18 [DOI] [PMC free article] [PubMed] [Google Scholar]