Figure 1.
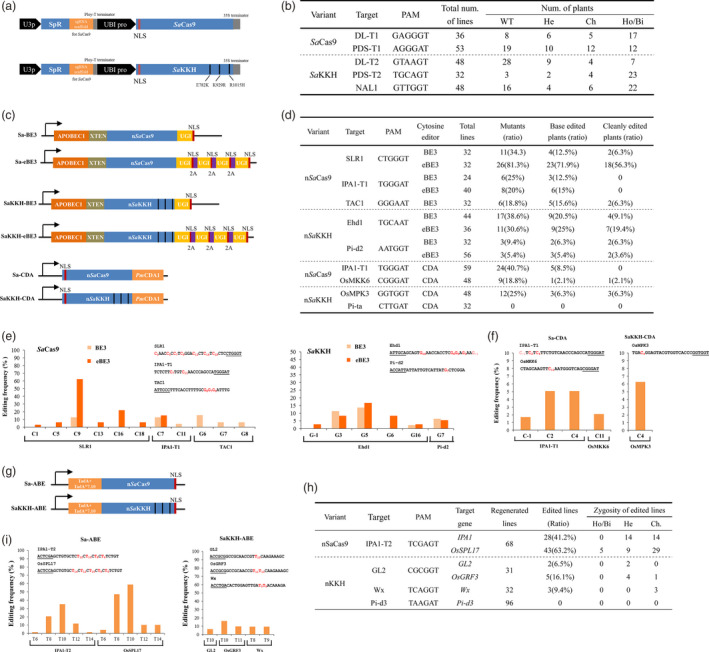
Rice genome editing generated by SaCas9 toolset. (a) Schematic illustration of the sgRNA and Cas9 expression cassettes of plant SaCas9 systems. To express sgRNA, a 21 bp protospacer sequence was inserted downstream of the rice U3 promoter (U3p) to replace the spectinomycin resistance gene (SpR). A maize ubiquitin promoter (UBI pro) was used to express SaCas9 or Sa KKH. (b) Mutations induced by SaCas9 and Sa KKH in regenerated rice plants analysed by Hi‐TOM assay with a 5% threshold (http://www.hi-tom.net/hi-tom). WT, wild‐type sequence in the target region; He, heterozygous mutation; Ch, chimeric mutation; Ho/Bi, homozygous or biallelic mutation. (c) Schematic illustration of SaCas9‐CBEs. (d) Cytosine editing induced by SaCas9 base editors in regenerated rice plants. The regenerated plants with exclusive C‐to‐T base conversions were considered as cleanly edited plants. (e) Frequencies of the base editing induced by SaCas9‐BE3 base editors at different C(G)s in the target sequence. The PAM sequence is underlined, and the targeted bases and positions in the protospacer are labeled in red. Sa‐eBE3 was not tested at the TAC1 target. (f) Frequencies of the base conversions induced by SaCas9‐CDA base editors at different C(G)s in the target sequence in regenerated plant populations. (g) Schematic illustration of SaCas9‐ABE base editors. (h) Adenine editing induced by SaCas9 base editors in regenerated rice plants. (i) Frequencies of the A‐to‐G conversion induced by SaCas9 ABEs at different A(T)s in the target sequence.
