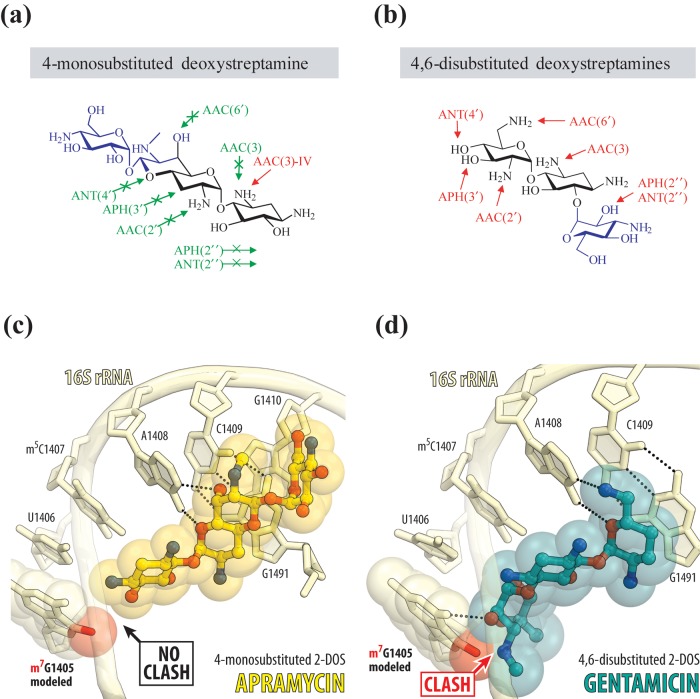
Figure 1.
Structural rationale for the activity of apramycin in the presence of aminoglycoside resistance determinants. The chemical structures of monosubstituted (a) versus disubstituted (b) deoxystreptamine antibiotics indicating the reactive groups that are modified by acetyltransferases (AACs), phosphotransferases (APHs) and nucleotidyltransferases (ANTs). Molecular modelling of the RMTase-catalysed N7-methylation of G1405 (red sphere) onto the crystal structure of ribosome-bound apramycin (PDB entry 4AQY) reveals no clash of the methyl group with the 4-monosubstituted 2-DOS apramycin (c). Molecular modelling of the G1405 methylation onto the crystal structure of ribosome-bound gentamicin (PDB entry 4V53) reveals considerable clash with ring III of 4,6-disubstituted 2-DOS (d). Nucleotides of the 16S rRNA are shown in pale yellow, and apramycin and gentamicin are shown in yellow and teal, respectively. The E. coli nucleotide numbering is used throughout.