Figure 2.
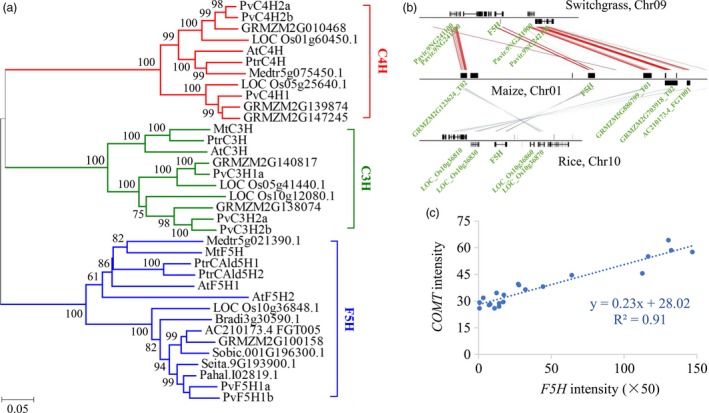
Molecular characterization of PvF5H. (a) Phylogenetic analysis of the three P450s (C4H, C3H and F5H) in the lignin biosynthetic pathway. A maximum likelihood tree was constructed in PhyML version 3.0 on the basis of multiple alignments of the deduced protein sequences from switchgrass, maize, sorghum, rice, Brachypodium distachyon, Arabidopsis thaliana, Medicago truncatula and Populus trichocarpa. Sequence data from this article can be found in Phytozome and/or Genbank under the following accession numbers: switchgrass Pavir.9NG241700.1 (PvF5H1a), Pavir.9KG138400.1 (PvF5H1b), Pavir.Fb01856.1 (PvC4H1), Pavir.5KG602000.1 (PvC4H2a), Pavir.5NG607400.1 (PvC4H2b), Pavir.3KG265800.1 (PvC3H1a), Pavir.5KG602000.1 (PvC3H2a), Pavir.5NG607400.1 (PvC3H2b); maize AC210173.4_FGT005 (F5H), GRMZM2G100158 (F5H), GRMZM2G139874 (C4H), GRMZM2G147245 (C4H), GRMZM2G010468 (C4H), GRMZM2G140817 (C3H), GRMZM2G138074 (C3H); sorghum Sobic.001G196300.1 (F5H); rice LOC_Os10g36848.1 (F5H), LOC_Os05g25640.1 (C4H), LOC_Os01g60450.1 (C4H), LOC_Os05g41440.1 (C3H), LOC_Os10g12080.1 (C3H); B. distachyon Bradi3g30590.1 (F5H); A. thaliana At4g36220 (AtF5H1), At5g04330 (AtF5H2), AT2G30490 (AtC4H), AT2G40890 (AtC3H); M. truncatula Medtr8g076290.1 (MtF5H), ABC59086.1 (MtC3H); and P. trichocarpa Potri.005G117500.1 (PtrCald5H1), Potri.007G016400.1(PtrCald5H2), Potri.013G157900.1 (PtrC4H), Potri.006G033300.1 (PtC3H). (b) Collinear relationships of F5H orthologs in genomes of switchgrass, maize and rice. A chromosomal region of PvF5H1a including 40‐kb flanking sequences were aligned with the corresponding orthologous sequences in maize (100 kb) and rice (40 kb). (c) Correlations between expression levels of F5H and COMT in different tissues and organs of switchgrass. The representative probesets of F5H (AP13ITG56842_at) and COMT (KanlowCTG00989_s_at) were retrieved from the switchgrass gene expression atlas. COMT, caffeic acid O‐methyltransferase.
