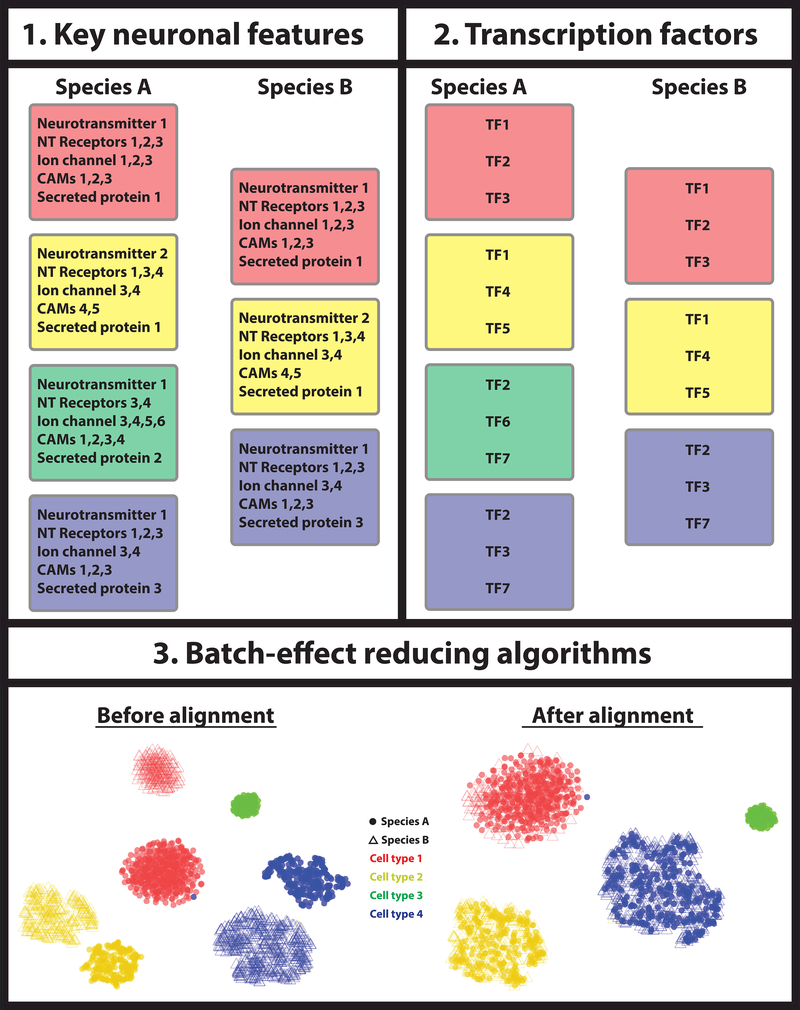
Figure 2: Identification of homologous cell types.
Three different approaches can be used to match homologous cell types between different species: 1) one can use key features that define each neuronal type, e.g. the expression of specific neurotransmitter, neurotransmitter receptors, ion channels, cell adhesion molecules (CAMs), secreted proteins etc. If the different species’ cell types express the same functional molecules, they are most likely homologous. 2) Alternatively, one can select a core regulatory complex of transcription factors and compare their expression between cell types of different species. Homologous cell types should express the same transcription factor fingerprint to regulate their phenotypic characteristics. 3) Finally, one could apply batch effect correction algorithms before comparing the different cell types between the two species, with the assumption that the effect of their independent evolution resembles a technical batch effect.