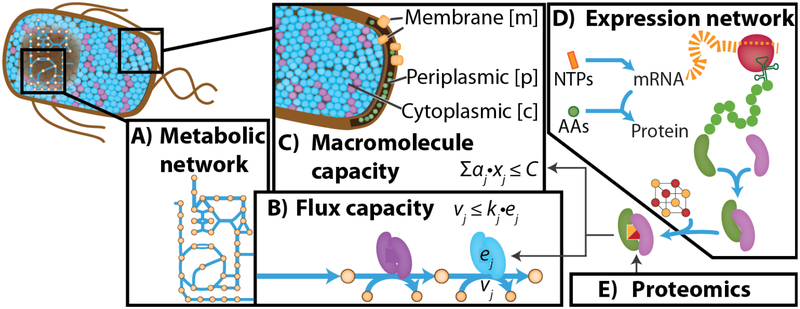
Fig 1 |.
Network models of metabolism and macromolecule expression. (A) a metabolic network is reconstructed from the microbe’s annotated genome. (B) Metabolic flux (vj) is constrained by catalytic efficiency (kj) and enzyme abundance (ej). This general form can be reformulated [34]. (C) Macromolecule (xi) capacity is constrained based on its physical properties (aj) such as molecular weight and a total cell capacity (C). Macromolecule abundance can be computed using a macromolecule expression network (D), which can vary in level of detail, or from proteomics data (E). To constrain total mRNA, RNA-Seq is used instead.