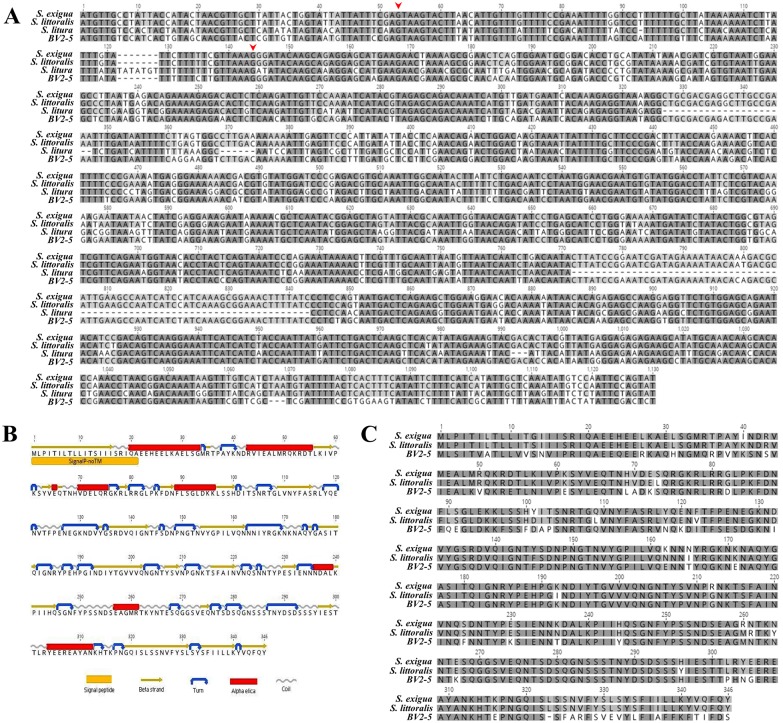
Fig 1. Sequence analysis.
DNA sequence alignment of gasmin homologues in 3 Spodoptera species and in CcBV (BV2-5) (the red arrowheads delimit the intronic region) (A), predicted secondary structure (B) and sequence conservation of Sl gasmin (C). Secondary structure prediction of Sl gasmin was carried out with the EMBOSS: Garnier algorithm; the Inter ProScan tool identified a potential signal-peptide. Bioinformatics analyses were performed using Geneious v6.1.6 (Biomatters, available from www.geneious.com) (A and C). DNA and protein alignments were performed using the Clustal W algorithm; black and grey shadings indicate identity and high conservation of amino acids, respectively.