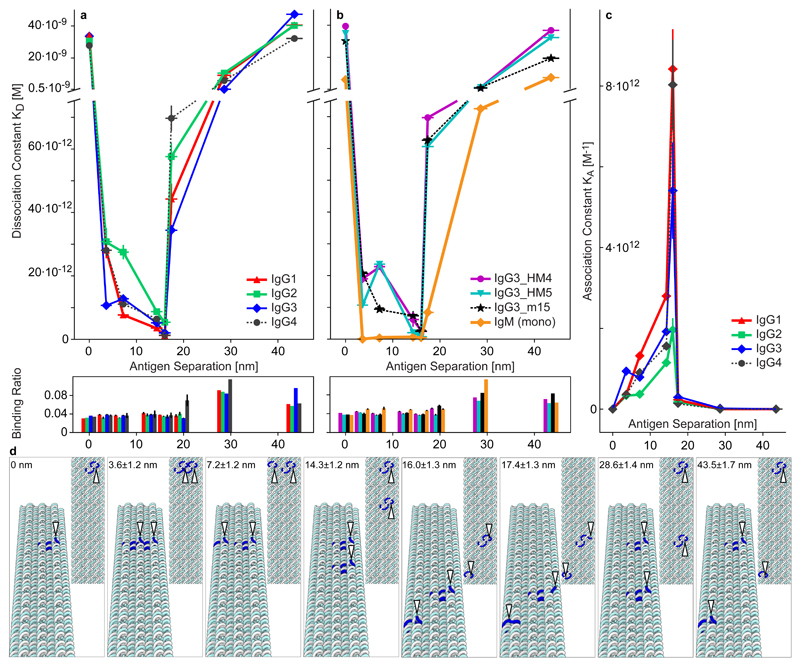
Fig. 2. Ab binding to precise antigen separations.
Human anti-NIP Abs binding to two NIP molecules patterned on the 18HB at varying separations. a-b Scatter graphs of KD and binding ratio (response units (RU):Abs / RU:structures, which correspond to average Abs per structure) for the various separations. See e and Fig S5 for exact structure designs. The “0” distance is just one displayed antigen. The y-axis (top parts) is the apparent binding affinity recorded and fitted with BIAcore t200 evaluation software. The amount of Ab 1 RU of structure can bind to is plotted in the bottom panels (binding ratio). a Data for all wt IgG subclasses. b Data for IgG3 hinge mutants and monomeric IgM. c The same data as in A plotted as association constants, KA (=1/KD), vs. separations for the IgG subclasses. A peak behaviour in the changes of KA was observed for all tested Abs, the bivalent binding becomes weaker at short (3-7 nm) and long (17 nm) distances, and this behaviour varies between different Ab subclasses. (n=3 or 4, detailed description is given in Supplementary Table 4; central values = average value; y error bars = standard error of mean, sometimes smaller than the markers; For samples without error bars n =1). Distance i.e. x-error bars are standard deviations from a uniform ±2 nm distribution added to a 3% standard deviation derived from structure fluctuations of the 18HB, see methods. d Zoomed in renders (top and perspective views) of the 18HB showing the locations of the NIP modified staple-oligos in blue, NIP locations highlighted with white arrowheads. The distances are shown as the design distance ± distance error calculated as described in methods.