Figure 1. ChP lipid, inflammation, and interferon (IFN) signatures.
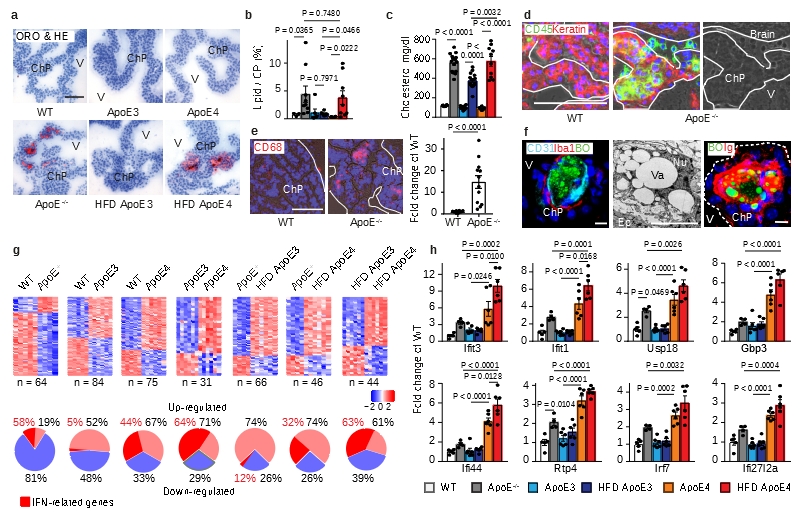
(a-b) ChP sections were stained with oil red o (ORO) for lipid (red) and hematoxylin (HE) for nuclei (blue) (a). Bar 100 μm; Representative images from b. ChPs and associated parenchyma per tissue area was quantified as described in Methods (b). WT (n=3 mice); ApoE-/- (n=9); ND ApoE3 (n=6); HFD ApoE3 (n=6); ND ApoE4 (n=6); HFD ApoE4 (n=9); (c) plasma cholesterol. WT (n=6); ApoE-/- (n=6); ApoE3 (n=14); ApoE3 HFD (n=17); ApoE4 (n=8); ApoE4 HFD (n=10) mice. (d) Epithelial cells were stained for cytokeratin (Keratin, red); leukocytes (CD45, green); and nuclei by DAPI (blue). Phase contrast delineates the ChP. Dashed line indicates the border of ChP and the ventricle. Bar 100 μm; (e) CD68+ areas were quantified as described in Methods. 12 sections from 4 WT; 12 sections from 4 ApoE-/- mice. (f) ChPs were stained for lipid by BODIPY (BO, green), endothelial cells (CD31, cyan), and macrophages (Iba-1, red) (left panel); TEM shows a single ChP macrophage-foam cell (middle panel); and lipid (BO, green) and immunoglobulin (Ig, red) (right panel). Bars 10 μm; (g) IFN-related genes in ChPs by microarray. Heat maps show two-group comparisons of ChPs. The percentages of up- or down-regulated genes were showed in the circle. The percentages of IFN-related gene in up- or down-regulated were marked as red. (h) ApoE4 isoform-dependent IFN signature expression in ChPs. WT (n=5 mice); ApoE-/- (n=4); ND ApoE3 (n=6); HFD ApoE3 (n=6); ND ApoE4 (n=6); HFD ApoE4 (n=6). Data in d,f are representative images from at least 3 biologically independent mouse samples. Data in b,c,e,h represent means ± SEM; Two-tailed Student´s t-test was applied to b,c,e; one-way ANOVA with Tukey’s test was applied to h; Abbreviations, ChP, choroid plexus; V, ventricle; Ep, epithelial cells; Nu, nucleus; Va, vacuole; TJ, tight junction; Gene names and statistics in supplementary Tabls.2-3.
