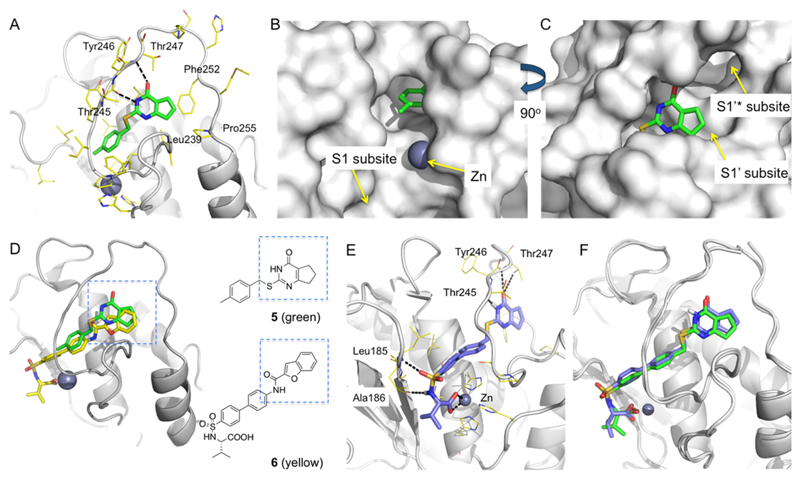
Figure 2.
Comparative structural analysis and design of Zn-chelating inhibitors. (A) X-ray crystallographic structure of MMP13·5 complex (PDB 4L19). Hydrogen bond interactions of 5 with the amide backbone units of Thr245 and Thr247 are represented in black dashed lines. Leu239, Phe252, and Pro255 form hydrophobic contacts with 5. (B) The 4-methylphenyl ring of 5 is oriented toward the Zn binding site and the MMP-13 S1 subsite. (C) The cyclopentyl ring of 5 occupies the S1′ subsite of MMP-13. (D) Superimposition of X-ray cocrystal structures of MMP-13·5 and MMP-13·6 (PDB 1ZTQ) complexes. (E) Docking model of the designed inhibitor (S)-10a in the MMP-13 active site (ligand–protein interactions are shown in black dashed lines). (F) Superimposition of the X-ray crystallographic (green) and model (blue) structures of MMP-13·(S)-10a complex. The X-ray crystallographic structure of MMP-13·(S)-10a was obtained at 1.60 Å (authors will release the atomic coordinates and experimental data upon article publication; PDB 5UWK). The docking structure was superimposed with the X-ray crystallographic structure by forming Cα atom pairs of MMP-13 using Schrödinger Maestro suites. The two ligand structures matched with 1.4 Å RMSD. Pymol was used to generate figures.