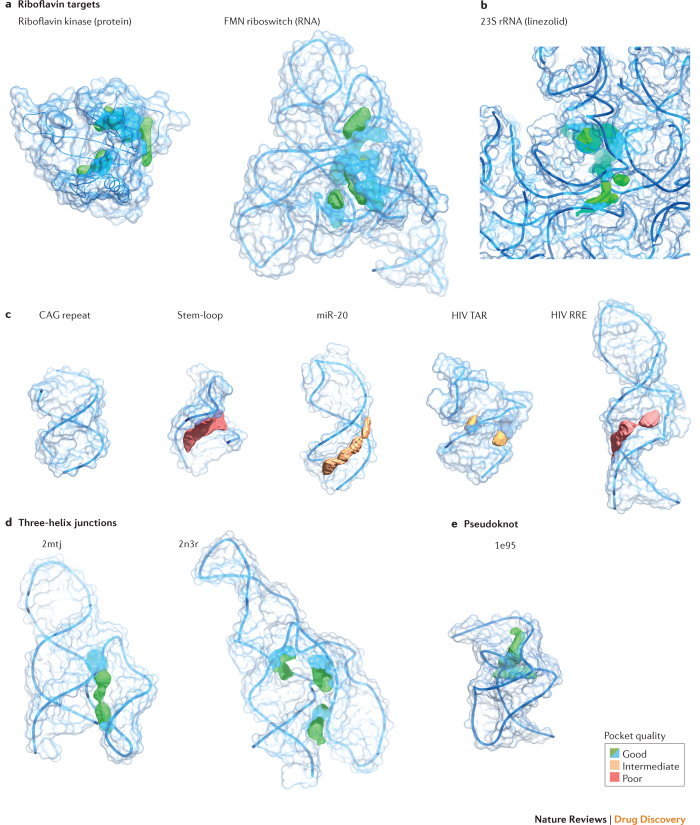
Figure 7. Pocket analysis of current and aspirational RNA targets.
Structures are coloured by pocket quality, with larger and more buried pockets ranking higher. a | Riboflavin binding pockets for protein (Protein Data Bank (PDB) identifier: 1nb9)69 and RNA (PDB ID: 3f4g)70 targets. Macromolecular targets are the same as those shown in Fig. 5. b | Pocket for linezolid in the Escherichia coli 23S ribosomal RNA (rRNA) (PDB ID: 3dll)29. c | Ligand-binding pockets in representative low-to-medium-complexity targets including a CAG helical repeat (PDB ID: 4j50)104, stem-loop (PDB ID: 1oq0)105, microRNA (miR; PDB ID: 2n7x)106 and HIV transactivation response (TAR) (PDB ID: 1qd3)107 and Rev response element (RRE) (PDB ID: 1i9f)108. d,e | Potential ligand-binding pockets in high complexity sites: three-helix junctions (PDB ID: 2mtj and 2n3r)82,83 and the simian retrovirus type 1 (SRV-1) pseudoknot (PDB ID: 1e95)85. Pocket qualities were calculated using Pocket-Finder81, as part of the MolSoft package. FMN, flavin mononucleotide.