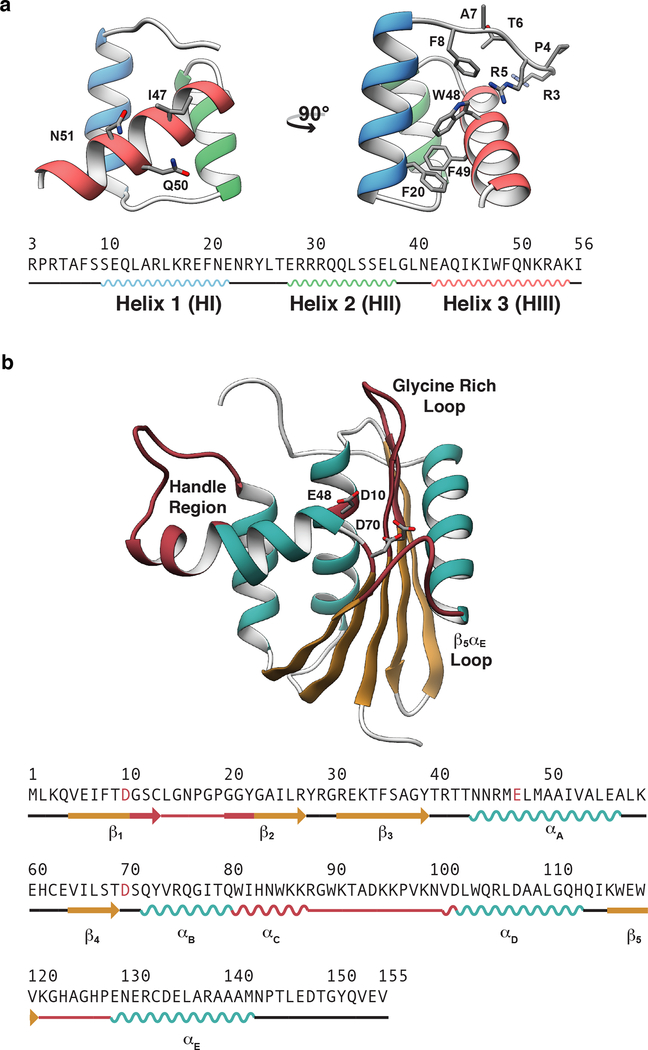
Figure 1. X-ray crystal structures of the engrailed homeodomain and ribonuclease H.
(a) The simple tertiary structure, small size, and ability to fold autonomously without either disulfide bonds or ligands have made homeodomains the subject of numerous investigations into the mechanisms of protein folding. Here, the crystal structure and sequence of the engrailed homeodomain are shown. On the front view (left), the DNA binding residues in HIII are represented as balls and sticks; on the side view (right), the DNA binding residues on the N-terminus as well as four aromatic residues within the hydrophobic core are shown as balls and sticks. (b) RNase H functions in numerous biological processes including inhibition of replication by removal of R-loops,51 removal of Okazaki fragments,52 synthesis of multicopy single-stranded DNA,53 and removal of misincorporated ribonucleotides.54 Here the crystal structure and sequence of ribonuclease H are shown with the β-sheet colored tan, α-helices cyan, and functional regions burgundy. Four functional regions of RNase H have been identified that are critical for RNase H to bind and hydrolyze RNA-DNA hybrids.55–57 These regions include αC and the loop between αC and αD (residues 81 to 101, referred to as the handle region), the loop between β1 and β2 (residues 11 – 22, referred to as the glycine rich loop), the loop between β5 and αE α/β (residues 121–127 referred to as the β5/αE loop), and the active site, which contains three conserved carboxylate residues (Asp10, Glu48, and Asp70) that coordinate divalent cations that are required for catalysis.