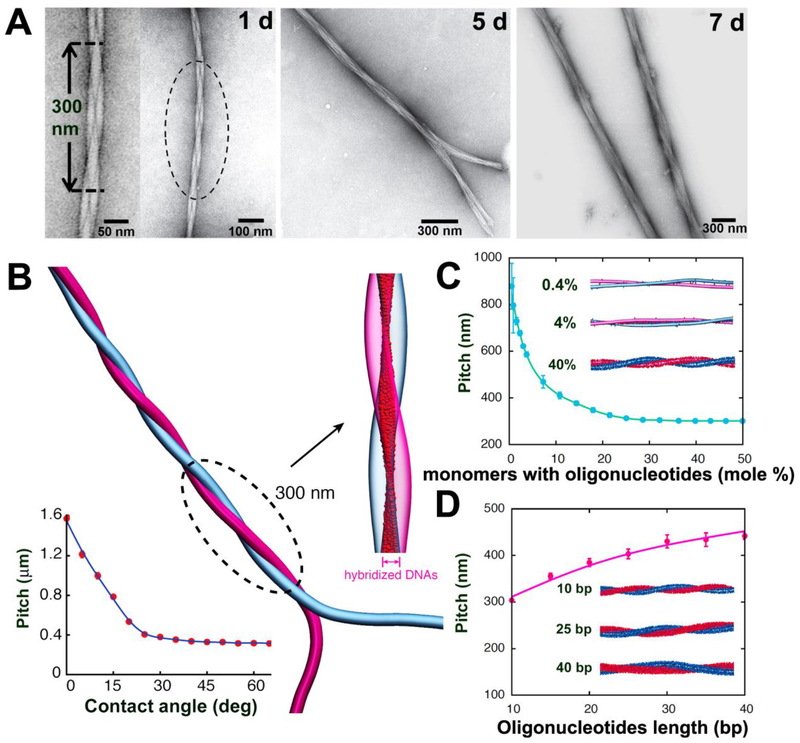
Figure 2. Programming the growth of intertwined bundles of fibers.
(A) TEM images after mixing complementary DNA- and PNA-terminated peptide amphiphiles show the time-dependent evolution of twisted bundles over 24 hours, 5 days, and 7 days. (B) Simulation snapshot of two intertwined complementary fibers. The intertwining pitch saturates for most initial contact angles (bottom left). Hybridized DNA-PNA pairs between the two fibers (magnified view) form a twisted ribbon pattern. (C) Dependence of the pitch on the fraction of monomers with oligonucleotides. Simulation snapshots shown for systems with 0.4%, 4%, and 40% oligonucleotides-modified monomers. (D) Dependence of the pitch on the oligonucleotides length. Simulation snapshots shown for duplexes with 10, 25, and 40 DNA-PNA base pairs (bp).