Figure 1. Distinct transcriptomic profiles in Shank2−/− brains at P14 and P25.
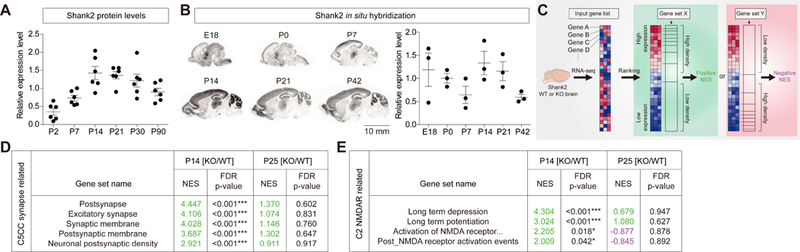
(A) Levels of Shank2 proteins in whole mouse brains increase during postnatal (P) days, reaching a plateau at ~P14. For quantification, Shank2 signals at each developmental stages were normalized to those of α-tubulin and also to the average values of each age groups. (n = 6 male mice for each stage). (B) Levels of Shank2 mRNAs in the mouse brain during development, revealed by in situ hybridization of sagittal sections. The limited correlation between Shank2 protein and mRNA levels might be attributable to that the transcripts are differentially translated or that the in situ images do not cover the whole brain. For quantification, Shank2 signals normalized to the brain area at each developmental stages were normalized to the average values of each age groups. (n = 3 male mice for each stage). Scale bar, 10 mm. (C) A schematic of gene set enrichment analysis (GSEA; software.broadinstitute.org/gsea/). The direction (positive/negative) and amplitude of enrichments reflect the extent to which the genes in a target gene set falls at the top (positive) or bottom (negative) of the ranked input gene list. (D) GSEA using KO/WT-P14 and KO/WT-P25 gene lists from Shank2−/− mice and target gene sets in the C5-CC category (gene ontology_cellular component). NES, normalized enrichment score accounting for the degree of overrepresentation at the top or bottom of a ranked list; FDR, false discovery rate. Positive and negative NES values are indicated in green and violet, respectively. (n = 6 mice for WT and KO at P14 and P25, ***FDR < 0.001; see Table S4 for details). (E) GSEA using KO/WT-P14 and KO/WT-P25 gene lists from Shank2−/− mice and NMDAR-related gene sets in the C2-all category. ‘Activation of NMDA receptor...’ indicates ‘Activation of NMDA receptor upon glutamate binding and postsynaptic event’. (n = 6 mice for WT and KO at P14 and P25, *FDR < 0.05, ***FDR < 0.001; see Table S3 for details).
