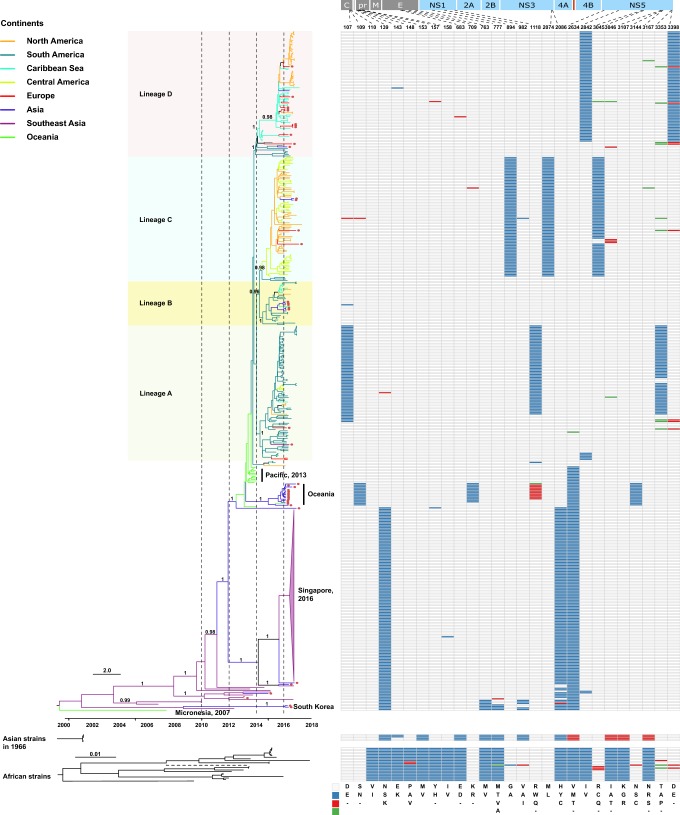
Fig. 1.
Phylogeny and molecular characterization of the Asian lineage of ZIKV. Full-length ZIKV genomes (n = 448) derived from the Asian lineage were assembled and a phylogenetic analysis using BEAST 1.8.4 was performed (Drummond et al.2012). The GTR + Γ nucleotide model was applied to account for rate variation among sites, with four categories for the Γ distribution. A log-normal distributed relaxed molecular clock was employed and the Bayesian skyline coalescent was designated as the tree prior. A Markov chain Monte Carlo with one hundred million steps was run twice independently and the first 10% were removed as burn-in. The posterior probability distribution of the parameters and trees from the two independent runs was combined using LogCombiner embedded in BEAST 1.8.4. Viruses identified from the ZIKV African lineage and from Indonesia cases in 1966 were also downloaded and phylogenetically analyzed using Maximum Likelihood. In the African lineage, one strain, ArD142623 (GenBank No. KF3838120), had a long branch length, which is displayed using a dashed line. In the Asian lineage, tips with circles represent confirmed ZIKV imported cases. Line color represents the collection site of the virus and the classification of the countries into different regions was based on Wikipedia (https://www.wikipedia.org/). In the right panel, different colors represent different amino acids, which have been shown in the legend in the bottom right of the panel.