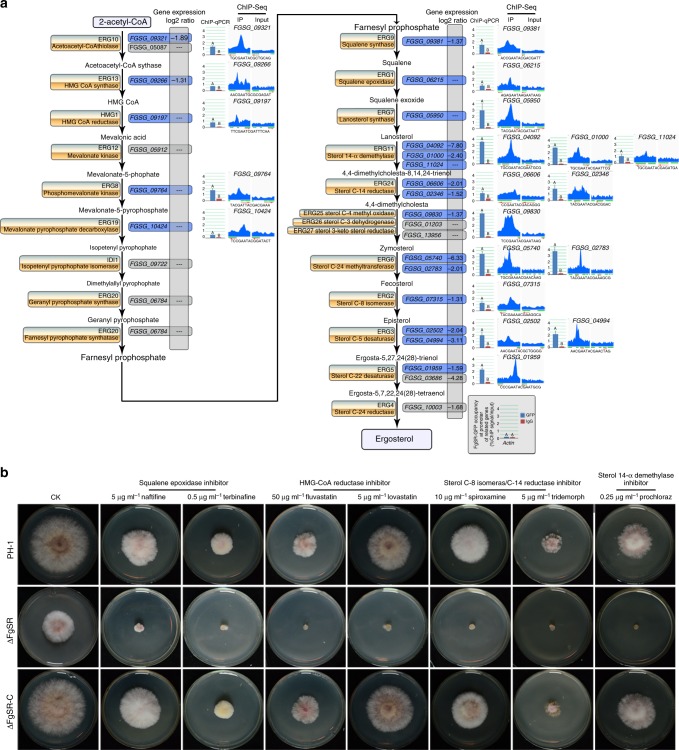
Fig. 6.
FgSR is a master regulator of ergosterol biosynthesis in F. graminearum. a ChIP-Seq and -qPCR assays showed that FgSR binds to the promoters of 20 ergosterol biosynthesis genes indicated in blue. RNA-Seq further confirmed that 14 out of the above 20 genes were down-regulated in the ΔFgSR mutant. FgACTIN was use as a negative control in the ChIP-qPCR assay. ChIP- and input-DNA samples were quantified by quantitative PCR assays with the primer pair A2 indicated in Fig. 2b, and rabbit IgG was used as a control. The red arrow indicates the gene direction from 5′ to 3′. The green bar represents the gene promoter region, which is covered during ChIP-Sequencing. “black dashed line” indicates no change in gene transcription determined by the RNA-Seq assay (Supplementary data 3). The sequences below the kurtosis graphs represent the cis-elements within the promoters of the 20 ergosterol pathway genes identified by MEME analysis. Values on the bars followed by the same letter are not significantly different at P = 0.01. b FgSR regulates F. graminearum sensitivity to sterol biosynthesis inhibitors (SBIs) targeted to different biosynthetic enzymes. A 5-mm mycelial plug of each strain was inoculated on PDA supplemented with 5 μg ml–1 naftifine, 0.5 μg ml–1 terbinafine, 5 μg ml–1 fluvastatin, 5 μg ml–1 lovastatin, 10 μg ml–1 spiroxamine, 5 μg ml–1 tridemorph, or 0.25 μg ml–1 prochloraz and then incubated at 25 °C for 3 days. As a control, each strain was cultured on PDA without supplementation