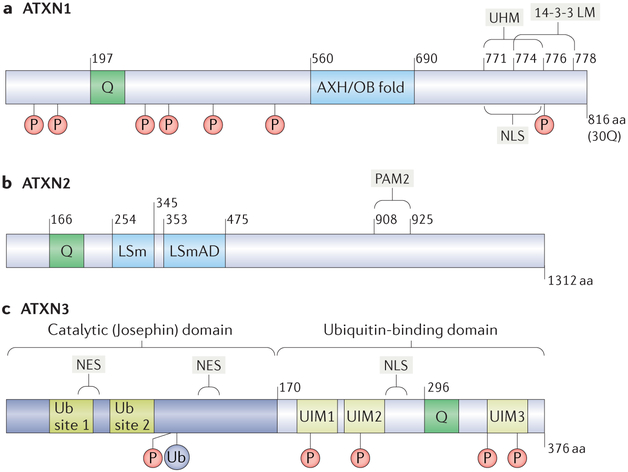
Figure 3 |. Functional motifs in ATXN1, ATXN2 and ATXN3.
Functional motifs are diagrammed for each polyglutamine (polyQ) spinocerebellar ataxia (SCA)-associated ataxin (ATXN) protein. The diagrams also show the polyQ regions (denoted as ‘Q’), as well as the phosphorylation sites (denoted as ‘P’) and ubiquitylation sites (denoted as ‘Ub’ to represent ubiquitin). a | ATXN1 with a 30Q polyQ region is shown. The ATXN1 HBP1 (AXH) domain and the U2AF homology motif (UHM) of ATXN1 are interaction motifs for capicua (CIC) and RBM17, respectively; the AXH domain itself forms an oligonucleotide/oligosaccharide-binding (OB) fold. ATXN1 also features a nuclear-localization signal (NLS) near the carboxyl terminus of the protein that facilitates its localization to the nucleus and a phosphorylation-dependent binding motif for the chaperone 14-3-3 (14-3-3 LM). b | ATXN2 interacts directly or indirectly with numerous proteins implicated in RNA metabolism. Its poly(A)-binding protein (PABP)-interacting motif PAM2 enables ATXN2 to interact with PABP and TAR DNA-binding protein 43 (TDP43). ATXN2 also features a like-Sm(LSm) motif and an LSm-associated domain (LSmAD). c | The deubiquitinase (DUB) ATXN3 has an N-terminal catalytic (Josephin) domain, which contains two Ub-binding sites and two nuclear export sites (NES), and a C-terminal Ub-binding domain bearing three Ub-interacting motifs (UIMs) and an NLS.