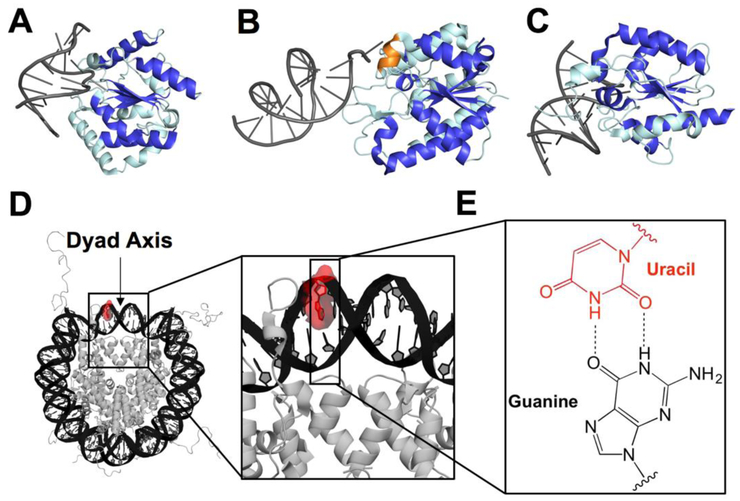
Fig. 1.
Graphical representation of co-crystal structures of uracil DNA glycosylases bound to duplex DNA, structure of an NCP, and U:G base pair. (A) UNG2 (PDB 1emh), (B) SMUG1 (PDB 1oe4), and (C) TDG (PDB 5hf7) bound to DNA with the central fold highlighted in royal blue and oriented using the β sheet motif. The helical wedge unique to SMUG1 is highlighted in orange. (D) Merged crystal structure of an NCP containing Widom 601 DNA (PDB 3lz0) and a histone octamer including histone N-terminal tails (PDB 1kx5). The U lesion is highlighted in red using PyMol and the dyad axis is indicated by an arrow. The DNA strand containing the U is numbered from the 5'-end (Widom 601 “i” strand), with the lesion located at position 75. (E) U:G wobble base pair used in this study as a mimic of C deamination.