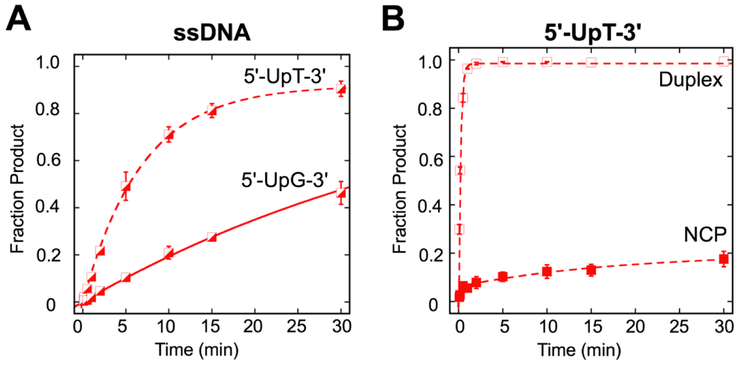
Fig. 3.
Single-turnover kinetics of SMUG1 activity in different sequence contexts. (A) Kinetic time courses were used to evaluate the excision of U in ssDNA substrates in 5'-UpT-3' (dashed line) and 5'-UpG -3' (solid line) sequence contexts. Reactions consisted of 20 nM DNA substrate, 640 nM SMUG1 in Buffer TNK. Data were fit to a single exponential equation. Error bars represent standard error (n=2 for 5'-UpT-3'; n=3 for 5'-UpG-3'). The observed rates and fraction product are kobs=0.14 ± 0.01 min−1, 0.92 (5'-UpT-3') and kobs=0.02 ± 0.01 min−1, 1.0 (5'-UpG-3'). (B) Kinetic time courses were used to evaluate the excision of U in duplex and NCP in a 5'-UpT/ApG-3' sequence context. Reactions consisted of 20 nM DNA substrate, 640 nM SMUG1 in buffer described in Materials and Methods. Data were fit to a single exponential equation (duplex), or double exponential equation (NCP). Error bars represent standard error (n=2 for duplex; n=6 NCP). Open squares ( ), duplex; filled squares (
), duplex; filled squares ( ), NCP. The observed rates (fraction product) are kobs=4.4 ±0.1 min−1 (0.99) (duplex) and kobs-fast=4.0 ± 0.9 min−1 (0.06), kobs-slow = 0.05 ± 0.02 min−11 (0.14) (NCP).
), NCP. The observed rates (fraction product) are kobs=4.4 ±0.1 min−1 (0.99) (duplex) and kobs-fast=4.0 ± 0.9 min−1 (0.06), kobs-slow = 0.05 ± 0.02 min−11 (0.14) (NCP).