Extended Data Figure 7. Analysis of histone acetylation, and ATAC-Seq overview and specific gene loci.
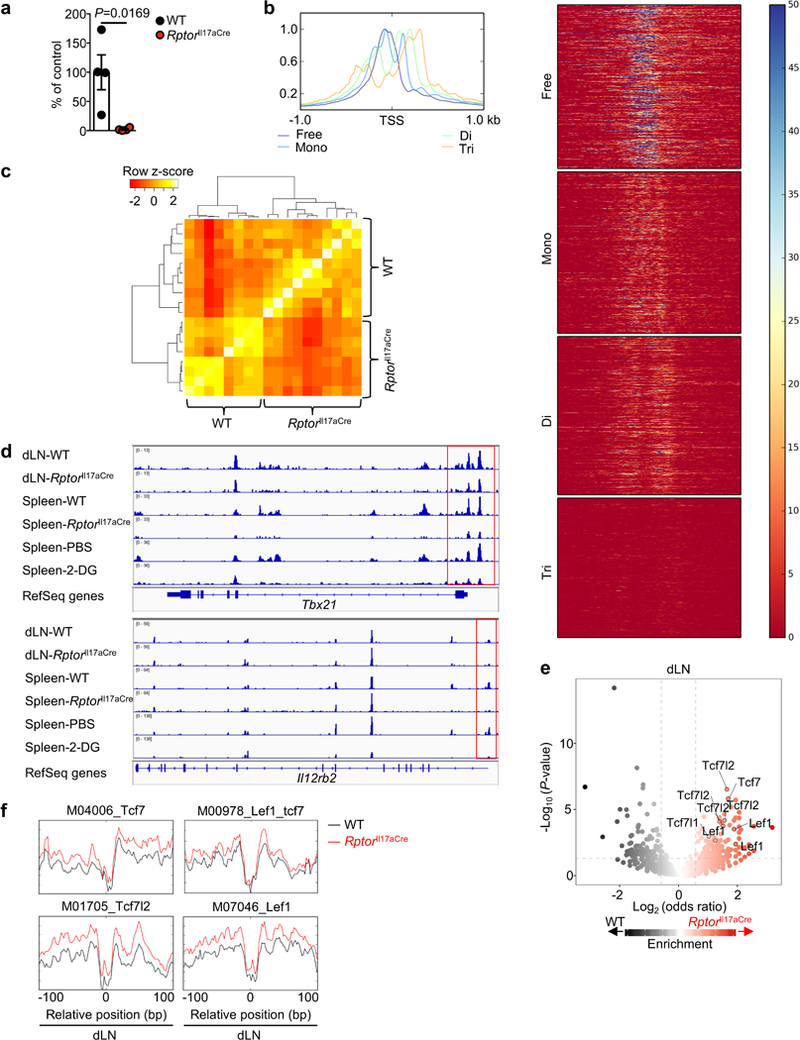
a, ChIP qRT-PCR of pan-acetyl histone bound to the Ifng promoter of WT or RptorIl17aCre (R26ReYFP) YFP+ cells from draining lymph nodes (dLN) (n = 4 per genotype). b, c, WT and RptorIl17aCre (R26ReYFP) mice were immunized with MOG, and YFP+ cells from dLN and spleen at day 9 post-immunization were analyzed by ATAC-Seq. b, Density plot and heat maps of a representative individual ATAC-Seq sample, demonstrating separation into different fragment lengths implicating nucleosome-free, mono-nucleosome, di-nucleosome, tri-nucleosome patterns, consistent with Buenrostro et al. c, Correlation plot of nucleosome-free fragments. d, Nucleosome-free ATAC-Seq tracks at the Tbx21 and Il12rb2 gene loci, with immediate promoter regions indicated by red boxes. e, Summary of ATAC-Seq motif enrichment data showing Log2 (odds ratio) and Log10 (Fisher P-value) of cells from dLN. f, Tn5 insert sites from ATAC-Seq analysis from dLN were aligned to motifs for transcription factors from the TRANSFAC database, and the binding profiles of selected TCF-LEF family transcription factors are shown. Data are means ± s.e.m and representative of three independent experiments (a), or from one experiment (b–f). Student’s t-test (two-sided; parametric) was used in a to determine statistical significance.
