Fig. 3.
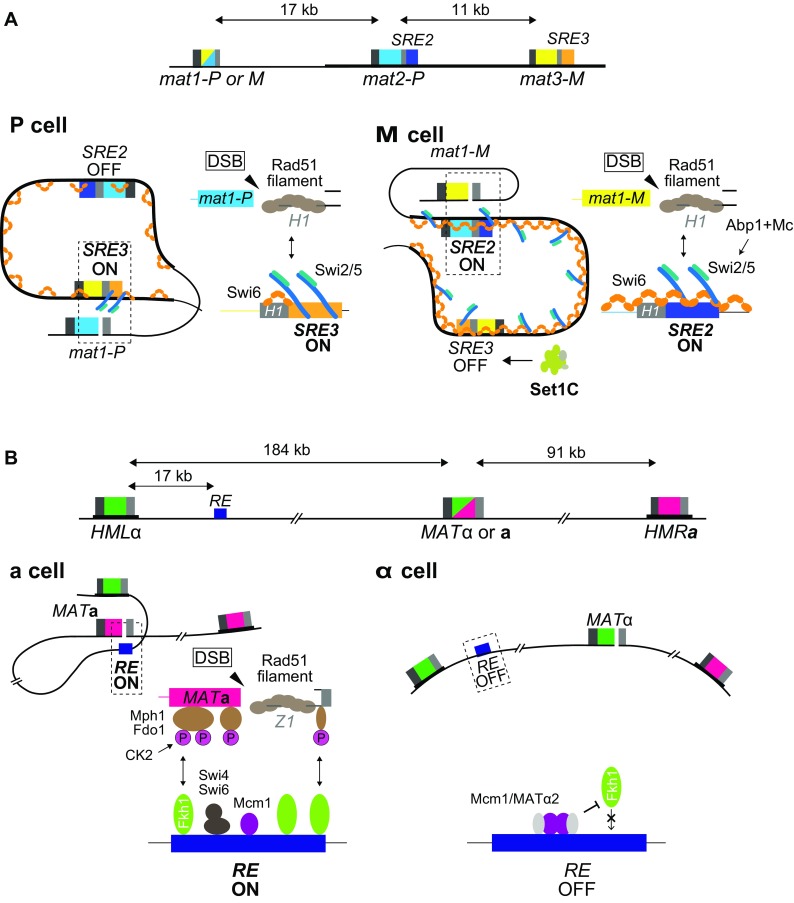
Choice of recombination enhancers in S. pombe and S. cerevisiae. aSchizosaccharomyces pombe SRE2 and SRE3 enhancers. In P cells, the Swi2/Swi5 complex is exclusively associated with the recombination enhancer SRE3; this facilitates Rad51-mediated strand invasion at the H1 region of mat3 and thus promotes switching from mat1-P to mat1-M. In M cells, the Swi2/Swi5 complex is more broadly associated with the entire heterochromatic domain, this association requires the chromodomain protein Swi6. Under these conditions, the SRE2 enhancer is preferred over SRE3 leading to switching from mat1-M to mat1-P. The use of SRE3 in M cells might be limited by Set1C. bSaccharomyces cerevisiae RE enhancer. In MATa cells, RE, bound by multiple copies of the Fkh1 protein, establishes a physical contact with the MAT locus through the interaction of Fkh1’s phospho-threonine binding domain with phosphorylated proteins located at or near the DSB; these proteins include Mph1 and Fdo1. RE binding at the site of the DSB shortens the distance between MATa and HMLα, favoring Rad51-mediated recombination between MAT and HMLα. In MATα cells, binding of Fkh1 to RE is disrupted by the repressor, Mcm1/Matα2; consequently, HMRa, closer to MAT, is then preferred
