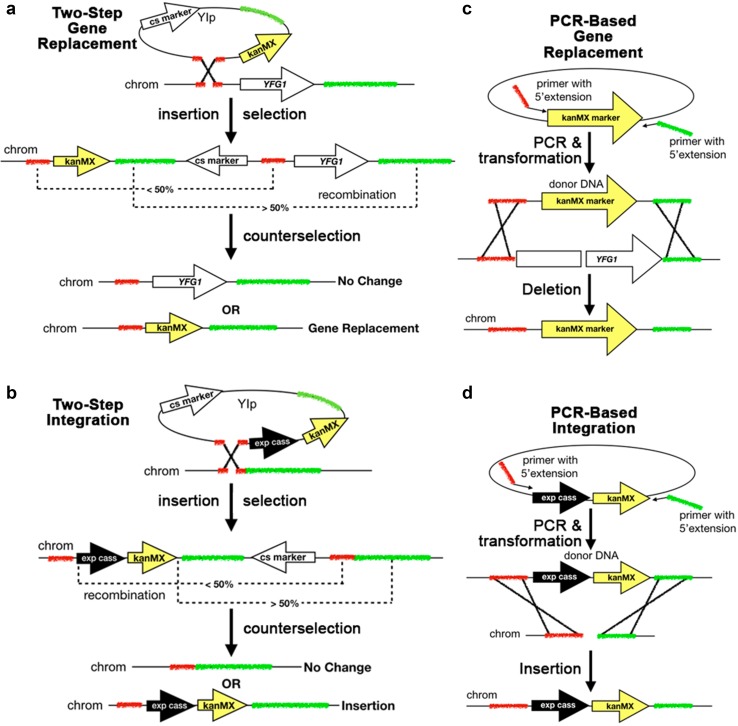
Fig. 2.
Gene editing approaches to make query mutant strains for GI screens YIp- or PCR-based methods for gene editing introduce donor DNA that serves as a template for homology-directed repair (HDR) when a spontaneous DNA double-strand break (DSB) occurs in the target locus denoted by your favorite gene yfg1∆. a, b YIp-based gene editing is performed in two steps. In the first step, the YIp linearized at a short cloned region of homology is integrated into the homologous chromosome region (red). The integrated construct generates direct repeats, which can spontaneously recombine and “pop-out” the construct. However, recombination is more likely to occur at the longer repeat region (green), resulting in stable integration of the construct. Recombinants are obtained by counterselection. a This Yip is designed to delete YFG1 and replace it with the kanR (kanMX) marker. b This YIp is designed to integrate an expression cassette (exp cass) in an intergenic chromosomal target. c, d PCR-based gene editing uses plasmid cassettes that contain a selectable marker. The target sequences are encoded in 40–60 nts of the PCR primer at 5′ overhangs (red and green). The primers also have ~ 20 nts of homology to the plasmid backbone flanking the cassettes at their 3′ ends (arrows). The same gene-specific primers can be used to insert various types of cassettes at the chromosomal target (red and green). c PCR-based gene deletion/replacement with a selectable marker. d PCR-based insertion of a marked expression cassette. kanMX: G418-resistance marker; YFG1: wild-type your favorite gene; X: homologous recombination event, chromI: chromosomal locus; exp cass: expression cassette, cs marker: counterselectable marker