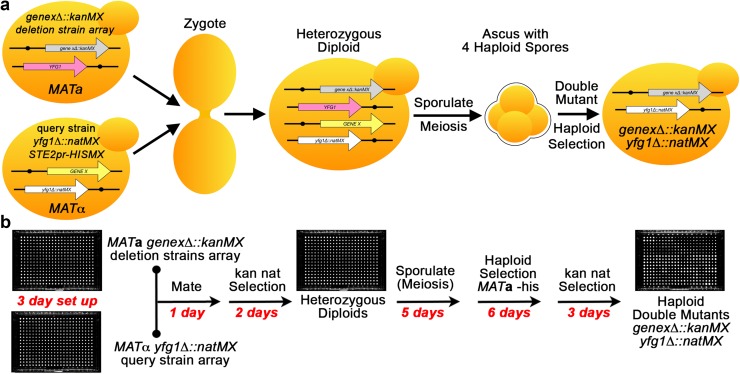
Fig. 3.
GI screens using SGA. a Schematic of the DNA molecules and manipulations resulting in the generation of a double mutant by SGA. b The SGA workflow starts with two agar plates of arrayed colonies. Each colony on the arrayed deletion collection plate is a different deletion strain marked with the same selection marker and in the same mating type. The query deletion strain, denoted by your favorite gene yfg1∆, is arrayed on a second plate and carries a different selection marker and has the opposite mating type. Colonies from the two parent plates are replica pinned onto the same plate to mix two mutants and allow them to mate and form a heterozygous diploid zygote. The diploids are replica pinned onto sporulation media. The resulting sporulation mix is plated on a series of selection media to obtain haploid strains carrying both parental deletions. Double mutants should be quadruplicated to 384 arrays for statistical measurements of growth. Plates are imaged and colony sizes are measured to determine if two genes interact. Red text indicates how many days are required for each step in the workflow. yfg1∆::natMX: query mutation of your favorite gene deleted and replaced with nourseothricin-resistance marker, genex∆::natMX: haploid strain collection gene deletion replaced with G418-resistance marker, STE2pr-HISMX MATa: haploid selection marker that confers growth on media lacking histidine