Fig. 5.
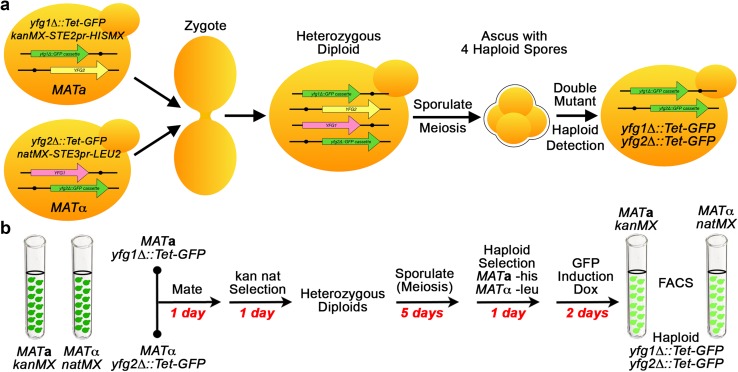
GI screens using green monster. a Schematic of the DNA molecules and manipulations resulting in the generation of a double mutant by Green Monster. b The Green Monster workflow starts with various strains from MATa and MATα haploid deletion collections in which the kanMX marker is replaced with a Tet-induced GFP expression cassette (denoted by your favorite gene yfg1∆ and yfg2∆). The MATa strains also carry a GMToolkit-a (kanMX-STE2pr-HISMX) and the MATα strains carry a GMToolkit-α (natMX-STE3pr-LEU2), both of which are derived from the SGA haploid selection reporter. These strains are randomly mated in bulk, and diploids are grown under double selection conditions. The resulting heterozygous diploids are sporulated and haploids of both mating types are selected. FACS analysis sorts cells that have twice the GFP intensity of the single mutants and, therefore, carry two deletions. The entire culture can be re-mated without FACS analysis to make mutants that carry 3, or more gene deletions. Red text indicates how many days are required for each step in the workflow. yfg1/2∆::Tet-GFP: query deletions of your favorite genes deleted and replaced with Tet-inducible GFP, kanMX-STE2pr-HISMX MATa: haploid selection marker that confers growth on media lacking histidine and marked with G418-resistance marker, natMX-STE3pr-LEU2 MATα: haploid selection marker that confers growth on media lacking leucine and marked with nourseothricin-resistance marker