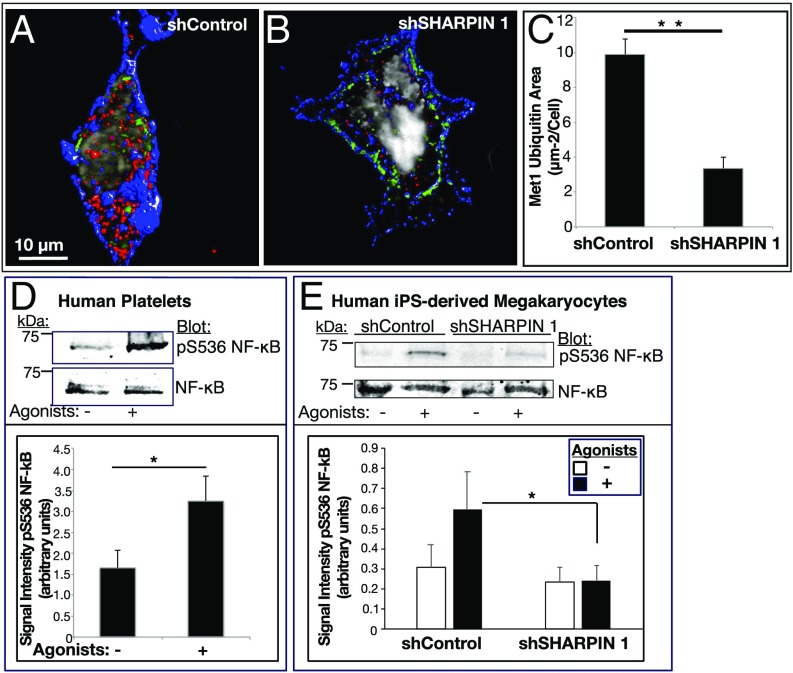
Fig. 6.
SHARPIN knockdown in megakaryocytes and platelets reduces Met1 ubiquitination and NF-κB pathway signaling. (A–C) Reconstructed immunofluorescence images of cells transduced with shControl RNA (A) or shSHARPIN 1 RNA (B). GFP-positive, shRNA expressing megakaryocytes were plated on fibrinogen and stained for Met1 ubiquitin (red), αIIb (blue), and DNA (gray). Images were acquired at 100× magnification. Met1 and αIIb are represented as isosurfaces. (C) Quantification of Met1 ubiquitination was carried out using Volocity image analysis software and is expressed as total Met1 ubiquitin area per cell. The data were collected from 3D projections of 13 optical slices of 0.2 μm each. A minimum of 20 cells were analyzed in each of three separate experiments (mean ± SEM; **P < 0.01). (D and E) Human platelets (D) or shControl- and shSHARPIN-treated megakaryocytes (E) were incubated for 15 min in the absence or presence of an agonist mixture (ADP, epinephrine and PAR1 agonist peptide as in Fig. 4). Cells were lysed and blots were probed with an antibody to phosphoserine-536 of the p65 (RelA) subunit of NF-κB and reprobed for total RelA. Quantification is represented as phosphoserine-536 RelA signal intensity normalized for total RelA NF-κB. (n = 4 in D; n = 8 in E; mean ± SEM; *P < 0.05).