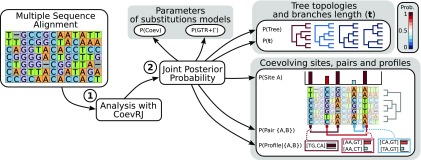
Fig. 1.
CoevRJ analysis flow. (1) A multiple sequence alignment containing nucleotides must be provided as input for CoevRJ. (2) After the analysis of the dataset, CoevRJ produces log files containing samples from the joint posterior distribution. These samples enable the estimation of the posterior probability of (i) the parameters of the evolutionary processes (GTR+ and Coev), (ii) the tree topologies and the branch lengths, and (iii) the pairs of sites and their profile. Further postanalyses with CoevRJ define the significance threshold for the coevolving pairs and provide easily readable summary statistics.