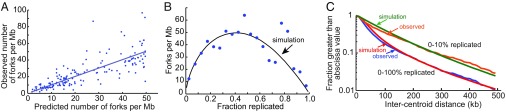
Fig. 2.
Simulation of DNA combing data. Simulations of S. pombe DNA replication based on the model described in Fig. 1 were performed with optimized firing parameters assuming an exponential distribution of the intervals between pre-RCs. The values of the parameters, optimized as described in Methods, were as follows: pre-RC density, 80 per Mb; rate of increase of firing probability per min, 0.022 events per min2 per pre-RC; maximum firing rate, 0.3 events per min per pre-RC; fork velocity, 2 kb/min. (A) Comparison of the observed density of replication forks with that predicted by the simulation. Each point represents a DNA molecule in the Kaykov and Nurse (21) dataset. The value on the ordinate is the observed density of forks in each molecule, and the value on the abscissa is the predicted density of forks for a molecule with the same percent replication. Slope of the regression line = 0.97; R2 = 0.63. (B) Comparison of the observed and predicted density of replication forks as a function of the fraction of the genome replicated. The observed fork densities (blue points) are the average observed values in windows of 5% replicated. The black line gives the fork densities predicted by the simulation. (C) Distributions of predicted and observed intercentroid distances [the distances between centers of replicated segments, referred to as interorigin distances in Kaykov and Nurse (21)]. The distributions are shown as semilog plots of the fraction of intercentroid distances greater than the value on the abscissa. Lower curves include 160 combed DNA molecules in the dataset of Kaykov and Nurse (21) with extents of replication ranging from 0 to 100%. Upper curves include a subset of molecules replicated from 0 to 10%.