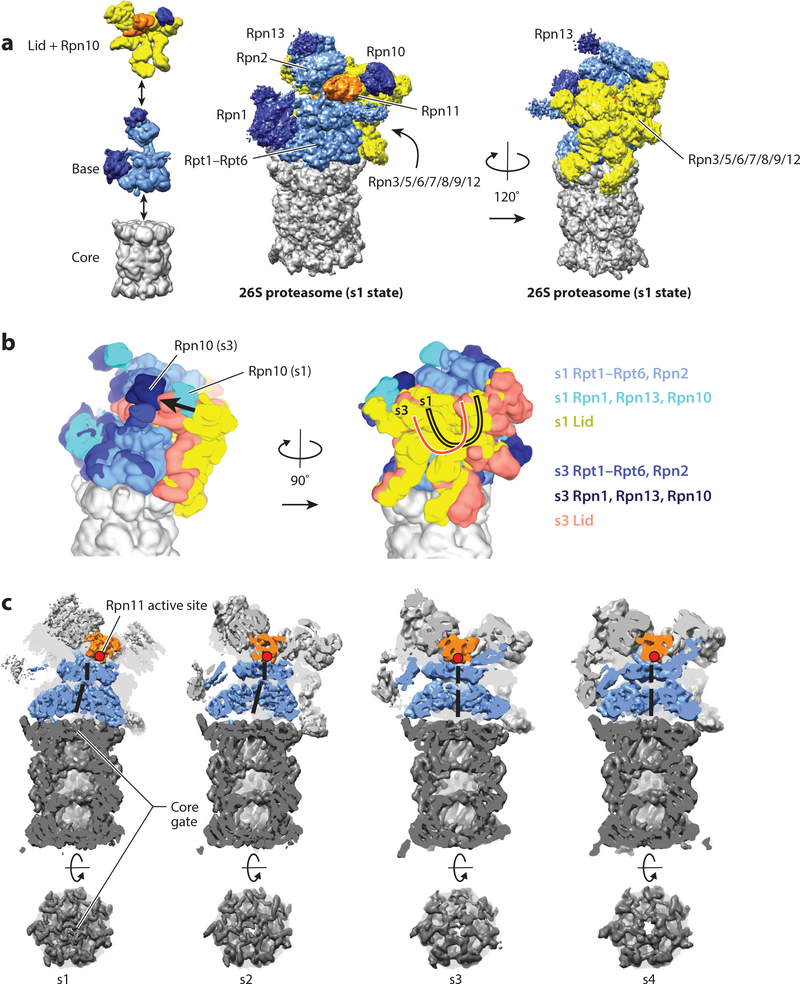
Figure 1.
Structure and conformational changes of the proteasome. (a) The 26S proteasome is composed of three subcomplexes: the core (gray); the base (with Rpn2 and the motor subunits Rpt1–Rpt6 in light blue and the ubiquitin-binding subunits Rpn1 and Rpn13 in dark blue); and the lid (with Rpn3, Rpn5, Rpn6, Rpn7, Rpn8, Rpn9, Rpn12, and Sem1 in yellow, and the DUB Rpn11 in orange). The ubiquitin receptor Rpn10 is shown together with the lid (dark blue). (Left) The three subcomplexes are depicted individually; (center and right) the entire 26S proteasome structure is shown (EMDB: 3534) (40). The center orientation allows a view of the entrance to the central pore and the Rpn11 active site, and the right orientation, rotated by 120°, emphasizes the lid subcomplex with its hand-shaped structure of the PCI (proteasome-CSN-initiation factor 3) domain-containing subunits. (b) Conformational switching of the 19S regulatory particle between the s1 state (EMDB: 3534) and the s3 state (EMDB: 3536) (40), with the core particles aligned. Shown are the views from the right and back of the proteasome relative to the center orientation in panel a. In the s1 conformer, the Rpt ring and Rpn2 are depicted in light blue; Rpn1, Rpn10, and Rpn13 in cyan; and the lid in yellow. In the s3 conformer, the Rpt ring and Rpn2 are depicted in medium blue; Rpn1, Rpn10, and Rpn13 in dark blue; and the lid in salmon. For both conformers, the core is shown in gray. During the transition from s1 to s3, the lid and Rpn10 rotate by ~30° relative to the Rpts. (c) Cutaway representations of the proteasome in the conformations s1–s4, emphasizing differences in the location of Rpn11; the width of the central processing channel; and the coaxial alignment of the N-ring, the AAA+ (ATPases associated with various cellular activities) ring, and the 20S core. The central channels through the N-ring and AAA+ ring are highlighted by a solid black line. The coaxial alignment is most pronounced in the s3 and s4 conformers, leading to the formation of a wide continuous channel for substrate translocation, with the Rpn11 active site (red dot) located directly above the entrance. Also shown are top-down views of the 20S core particle, emphasizing the changes in the 20S gate, which has the most density in the s1 state and the least in the s4 state.