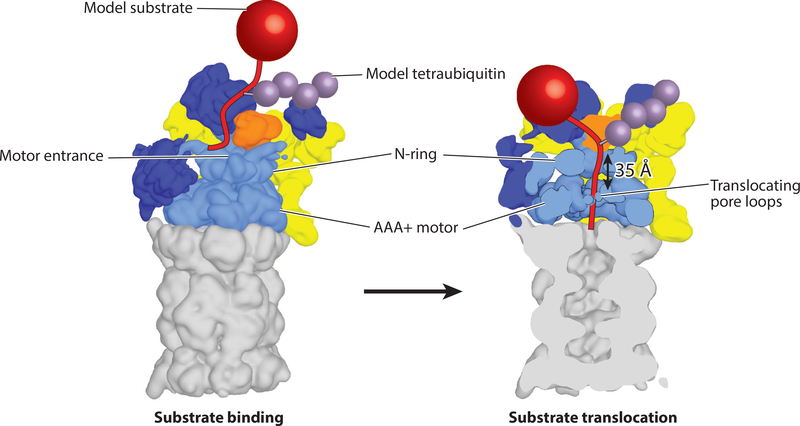
Figure 2.
Model for substrate engagement by the proteasome. (Left) Proteasome is shown in the s1 state (EMDB: 3534) with a model substrate (red) tethered through a tetraubiquitin chain (purple) near the presumed location of the Rpn10 ubiquitin-interacting motif. In this s1 state, Rpn11 (orange) is offset to the right, making the entrance to the central pore accessible for insertion of a substrate’s unstructured initiation region. (Right) Substrate engagement shifts the proteasome to the s3 or s4 state (s4 is shown; EMDB: 3537) (40) in which the N-ring and AAA+ (ATPases associated with various cellular activities) ring are coaxially aligned to facilitate substrate translocation and the 20S gate is open for polypeptide transfer into the internal degradation chamber. Rpn11 is located directly above the entrance to the central pore, where it acts as a gatekeeper and removes ubiquitin chains from substrates during translocation. The initiation region of the substrate must be long enough to bridge the gap between the N-ring and the pore loops of the Rpts.