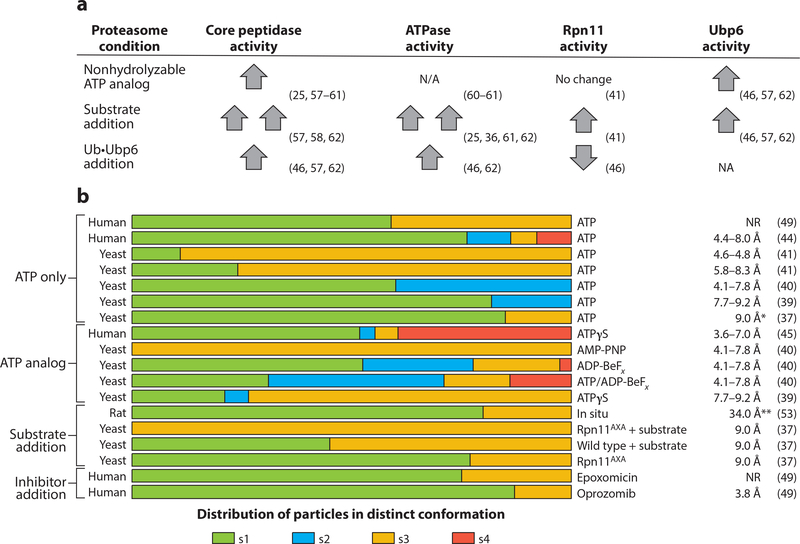
Figure 3.
Conformational landscape of the proteasome. (a) Qualitative comparison of how the presence of nonhydrolyzable ATP analogs, protein substrate, and ubiquitin-bound Ubp6 affects individual enzymatic activities of the proteasome. Up and down arrows indicate the stimulation and inhibition, respectively, of enzymatic activities of the proteasome compared with their basal activities; two up arrows indicate hyperstimulation. Basal activities are defined here for the proteasome in the presence of ATP and absence of substrate proteins. (b) Data from 17 electron microscopy data sets that directly compare the relative abundances of proteasome conformations under various conditions. The data sets are clustered according to the experimental conditions. Increased ATPase and core-peptidase activities appear to be correlated with greater abundance of s3 and s4 states. (Right) Resolutions of all structures obtained, representing the FSC value at 0.143. An asterisk indicates resolution was attained with an FSC value of 0.3; double asterisks indicate resolution was attained with an FSC value of 0.5. Abbreviations: FSC, Fourier shell correlation; NA, not applicable; NR, not reported; Ub, ubiquitin.