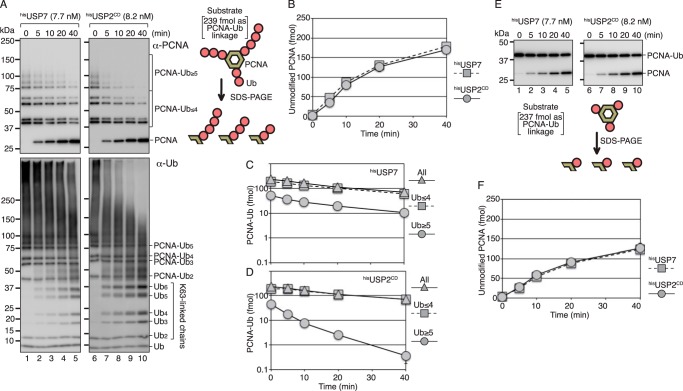
Figure 1.
Isopeptidase activity of USP7 and USP2 for polyubiquitinated and monoubiquitinated PCNA. A, the polyubiquitinated PCNA substrate (80 fmol as a trimer) consisted of 239 fmol of modified PCNA subunits and 1 fmol of unmodified PCNA subunits. The ubiquitinated PCNA contained 195 fmol of PCNA subunits with 1- to 4-mer ubiquitin chains (PCNA-Ub ≤4), and 44 fmol of PCNA subunits with ubiquitin chains longer than 5 mer (PCNA-Ub ≥5) (lanes 1 and 6). The substrate was incubated with hisUSP7 (7.7 nm) or hisUSP2CD (8.2 nm) for the indicated times. The reaction products were analyzed by Western blotting with an anti-PCNA mAb (upper panels) or an anti-ubiquitin mAb (lower panels). B–D, quantified data. Signal intensities of the indicated areas in (A) (PCNA, PCNA-Ub ≤4, and PCNA-Ub ≥5) were measured, and the relative amounts of PCNA (unmodified), PCNA-Ub ≤4, PCNA-Ub ≥5, and total modified PCNA were calculated. The averages of two independent experiments were plotted. In most cases, the error bars were smaller than the symbols. The amounts of unmodified PCNA generated in the reactions (B) and the amounts of remaining ubiquitinated PCNA in the reaction with hisUSP7 (C) or hisUSP2CD (D) are shown. E, the monoubiquitinated PCNA substrate (80 fmol as a trimer) consisting of 237 fmol of modified PCNA subunits and 3 fmol of unmodified PCNA subunits was incubated with hisUSP7 (7.7 nm) or hisUSP2CD (8.2 nm) for the indicated times. The reaction products were analyzed by Western blotting with an anti-PCNA mAb. F, quantified data. Signal intensities of PCNA and PCNA-Ub1 in (E) were measured, and the relative amounts of PCNA (unmodified) were calculated. The averages of two independent experiments were plotted. The error bars were smaller than the symbols.