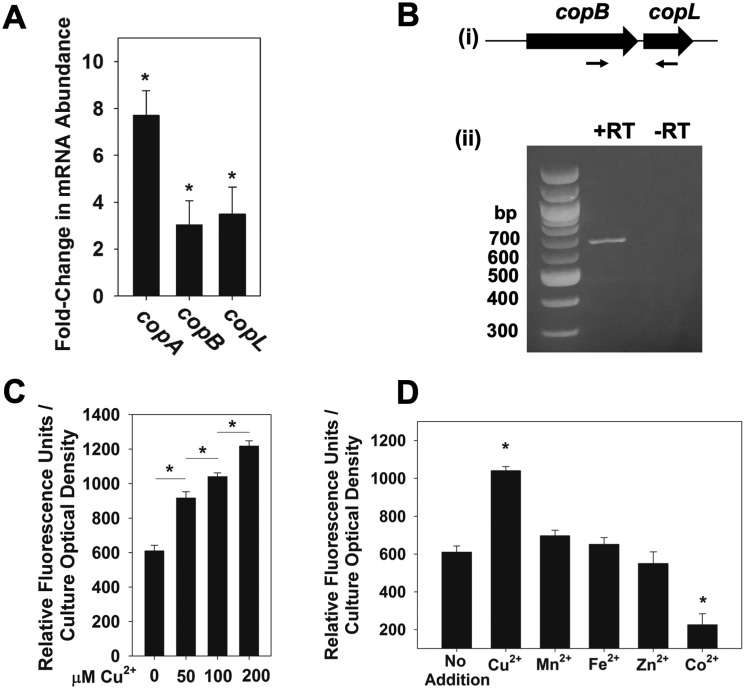
Figure 3.
copBL operon is up-regulated under copper stress. A, copA, copB, and copL genes are induced by Cu. The S. aureus WT (JMB1100) was cultured in chemically defined media containing 0 or 100 μm Cu. RNA was isolated; cDNA was generated, and the abundance of the copA, copB, and copL transcripts was quantified using qPCR. Data show fold-induction of genes of interest upon addition of Cu with respect to no Cu addition. Data represent the average of biological triplicates with errors presented as standard deviations. B, copB and copL genes are co-transcribed. RNA was isolated from the WT grown in chemically defined medium with 100 μm Cu and cDNA libraries generated. Panel i, schematic showing the primer pair (ZRT21 and ZRT24) used to detect the copBL transcript; expected size: 643 bp. Panel ii, agarose gel electrophoresis was used to detect the copBL amplicon generated using cDNA libraries as template DNA. A reaction without reverse transcriptase (−RT) was included as a control for genomic DNA contamination. C, transcriptional activity of copB increases in synchrony with Cu concentration. Transcriptional activity of the copB reporter was monitored in the S. aureus USA300 WT grown in chemically defined media containing 0, 50, 100, or 200 μm Cu. D, transcriptional activity of copB is specific to copper stress. Transcriptional activity of copB was monitored in the WT grown in chemically defined media containing 100 μm Cu2+, 100 μm Mn2+, 100 μm Fe2+, 100 μm Zn2+, or 50 μm Co2+. C and D, fluorescence data were standardized to culture optical density (A590 nm), and data represent the average of biological triplicates with errors presented as standard deviations. Student's t test were performed on the data, and * indicates p ≤ 0.05.