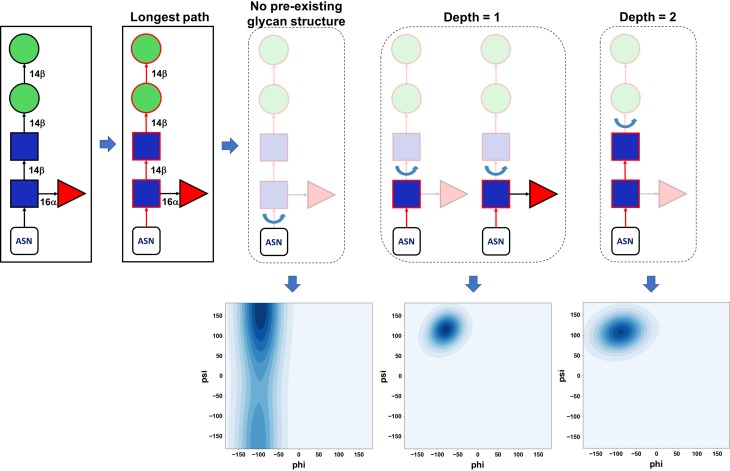
Fig. 6.
Rotatable glycosidic linkage. Graphical illustration of rotatable glycosidic linkage determination depending on the extent of a pre-existing glycan structure in a protein. After the longest path on the glycan sequence at each glycosylation site is identified (red), a rotatable glycosidic linkage is defined as a linkage between residues with and without coordinates on the longest path. If there are no coordinates only for the last glycans (i.e., the depth more than two in this case), they are generated simply by IC information in the CHARMM force field and has no rotation during the modeling step. Note that all glycosidic linkages in the pre-existing glycan structure are fixed. Random glycosidic torsion angles are generated based on glycosidic torsion angle populations from GFDB by using the kernel density estimation algorithm in a scikit-learn python module (Pedregosa et al. 2011).