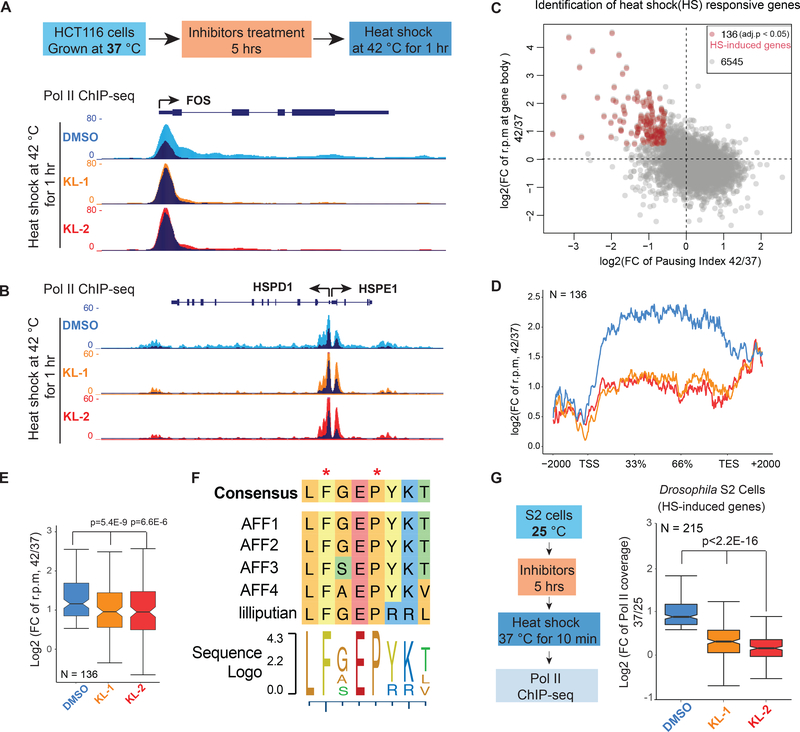
Figure 5. SEC inhibitors block transcription elongation in SEC-dependent rapid response models.
(A-B) Genome browser views of Pol II ChIP-seq at FOS, HSPD1 and HSPE1 after 1 hr heat shock of HCT-116 cells treated with vehicle (DMSO) or the indicated SEC inhibitors. HCT-116 cells were pretreated with inhibitors for 5 hr at 37 °C before exchanging medium with conditioned 42 °C medium. Dark and light colors indicate the 37 and 42 °C conditions, respectively.
(C) Genome-wide identification of heat shock-induced genes with fold change of reads per million (rpm) in gene bodies and pausing index. Red dots are the 136 heat shock-induced genes according to Pol II signals (fold change rpm in gene bodies > 1.5 and pausing index decreased > 1.5-fold in the DMSO treatment).
(D-E) Metagene plot of the 136 heat-shock induced genes shows attenuated induction with both inhibitors (D). Box plot analysis depicts the log2 fold changes (rpm) in gene bodies (E) (y axis) after heat shock with pretreatment of vehicle or SEC inhibitors.
(F) Sequence alignment of the CCNT1 interacting region in human AFF family proteins and the Drosophila homolog Lilliputian.
(G) Genome-wide analysis of SEC inhibitors effect on the heat shock response in Drosophila S2 cells (N=215). S2 cells treated with and without 20 μM SEC inhibitors were heat shocked 10 min at 37 °C before performing Pol II ChIP-seq. Statistical analysis was performed with the Wilcoxon test. See also Figure S5.