Figure 4.
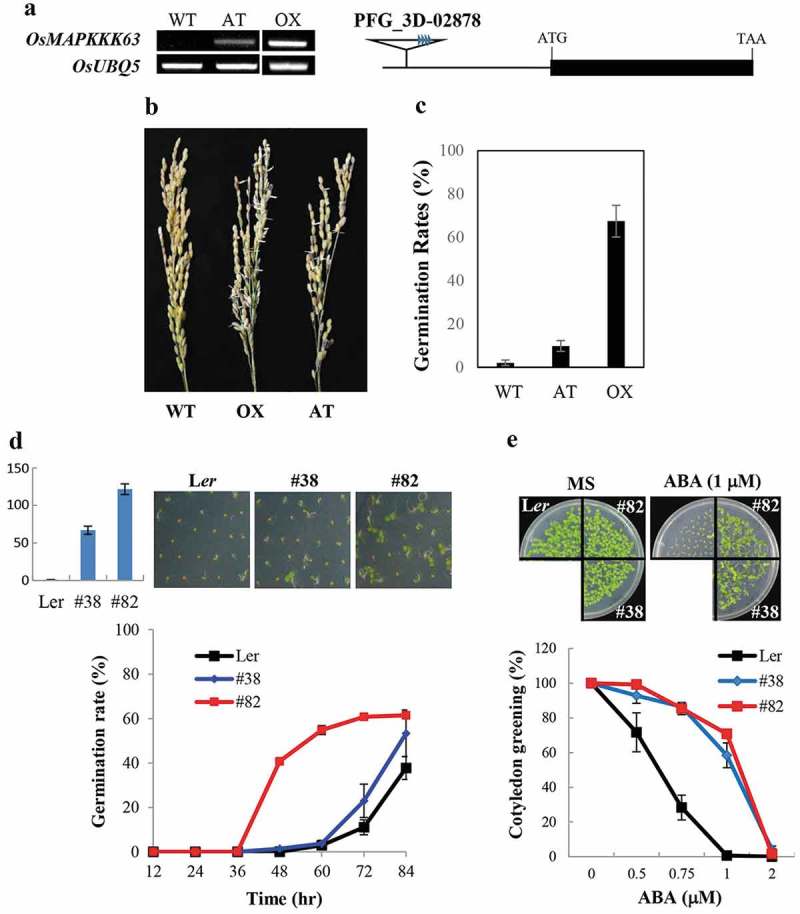
Seed dormancy of OsMAPKKK63 OX lines. (a) Left, OsMAPKKK63 expression levels in the transgenic rice plants determined by semiquantitative RT-PCR. AT, activation-tagged line. OX, OX line. Rice ubiquitin5 gene was used as a reference gene. Right, a schematic diagram of rice activation-tagged line (PFG_3D-02878). (b) Rice panicles harvested from the field-grown plants and kept in high humidity (99%) condition for two days. Please note that OX and AT line seeds sprouted. (c) Vivipary of OX and AT lines. Preharvest sprouting (i.e. germination of seeds in the live panicle of field-grown plants) is presented as germination rates. (d) Seed dormancy of Arabidopsis OX lines. Top left, Expression levels of OsMAPKKK63 in the transgenic lines determined by Real-Time RT-PCR. Top right, Photographs showing representative germinating seeds. Bottom, Germination rates of freshly harvested seeds. Seeds were plated without stratification and germination (radicle emergence) rates were scores at indicated time points. The numbers indicate line numbers, and small bars indicate standard errors. (e) ABA sensitivity of Arabidopsis OX lines during cotyledon greening/expansion stage. Top panels show seedlings grown for four days in MS medium or MS medium containing 1 μM ABA. Bottom, cotyledon greening efficiency. Seeds were germinate and grown for four days before counting seedling with green cotyledons. Bars indicate standard errors.
