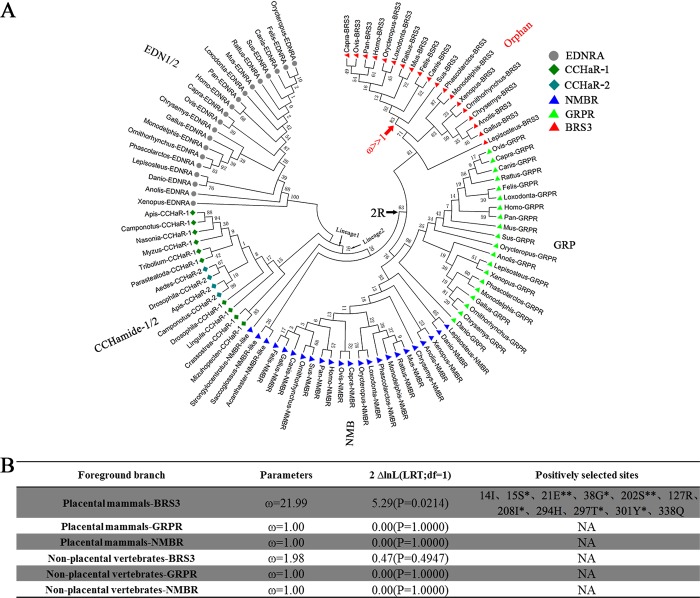
Fig 1. The evolution of the bombesin receptors.
(A) Phylogenetic tree of bombesin receptors. The ligands were shown in vicinity to their corresponding receptors. The ligand for BRS3 remained elusive thus far. The species of phylogenetic tree are as follows: Homo: Homo sapiens; Pan: Pan troglodytes; Mus: Mus musculus; Rattus: Rattus norvegicus; Sus: Sus scrofa; Capra: Capra hircus; Ovis: Ovis aries; Canis: Canis lupus familiaris; Felis: Felis catus; Orycteropus: Orycteropus afer; Loxodonta: Loxodonta africana; Phascolarctos: Phascolarctos cinereus; Monodelphis: Monodelphis domestica; Ornithorhynchus: Ornithorhynchus anatinus; Gallus: Gallus gallus; Anolis: Anolis carolinensis; Chrysemys: Chrysemys picta; Xenopus: Xenopus tropicalis; Lepisosteus: Lepisosteus oculatus; Danio: Danio rerio; Saccoglossus: Saccoglossus kowalevskii; Acanthaster: Acanthaster planci; Strongylocentrotus: Strongylocentrotus purpuratus; Apis: Apis mellifera; Nasonia: Nasonia vitripennis; Drosophila: Drosophila melanogaster; Aedes: Aedes aegypti; Tribolium: Tribolium castaneum; Camponotus: Camponotus floridanus; Parasteatoda: Parasteatoda tepidariorum; Myzus: Myzus persicae; Lingula: Lingula anatina; Crassostrea: Crassostrea virginica; Mizuhopecten: Mizuhopecten yessoensis. (B) The parameters and statistical significance of LRTs for each branch of BRS3, GRPR, and NMBR are given; the dN/dS ratio is calculated with the whole protein coding region; **, *, and no * indicate P values in excess of 0.99, 0.95, and 0.90, respectively. 2R, two rounds of whole-genome duplications; BRS3, bombesin receptor subtype-3; CCHaR-1, CCHamide-1 receptor; CCHaR-2, CCHamide-2 receptor; df, degree of freedom; EDNRA, endothelin receptor type A; GRP, gastrin-releasing peptide; GRPR, GRP receptor; LRT, likelihood ratio test; NA, not available; NMBR, neuromedin B receptor.