(
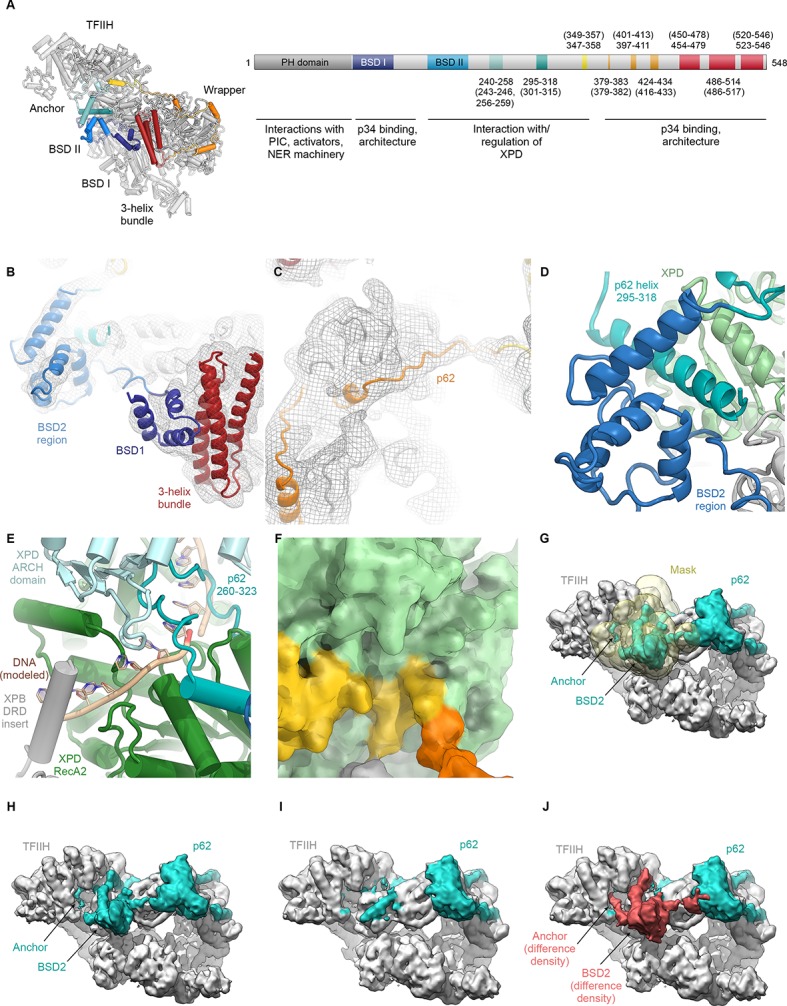
A) p62 architecture and sequence assignment. Fitting of domains (see
B), secondary structure prediction for α-helices (
Kelley et al., 2015), helical segments of the cryo-EM map, and density for large side chains were used for initial assignment of the trace of p62. The final model shows good agreement of observed helical segments (residue numbers indicated) and secondary structure prediction for α-helices (residue numbers given in brackets). (
B) Fitting of p62 BSD1 (PDB ID 2DII; blue) into the TFIIH cryo-EM map. Due to flexibility, the density in this region is weak and was low-pass filtered to 6 Å for interpretation and fitting. In agreement with previous results (
Schilbach et al., 2017), this domain interacts with the p62 C-terminal three-helix bundle (purple), which in turn sits on the eZnF domain of p44 (
Figure 1—figure supplement 6J), thereby explaining the importance of the latter region for functional TFIIH (
Tremeau-Bravard et al., 2001). (
C) Extended segments of the p62 chain were interpreted using the low-pass filtered map as described in (
B) to maintain continuity of the cryo-EM density. (
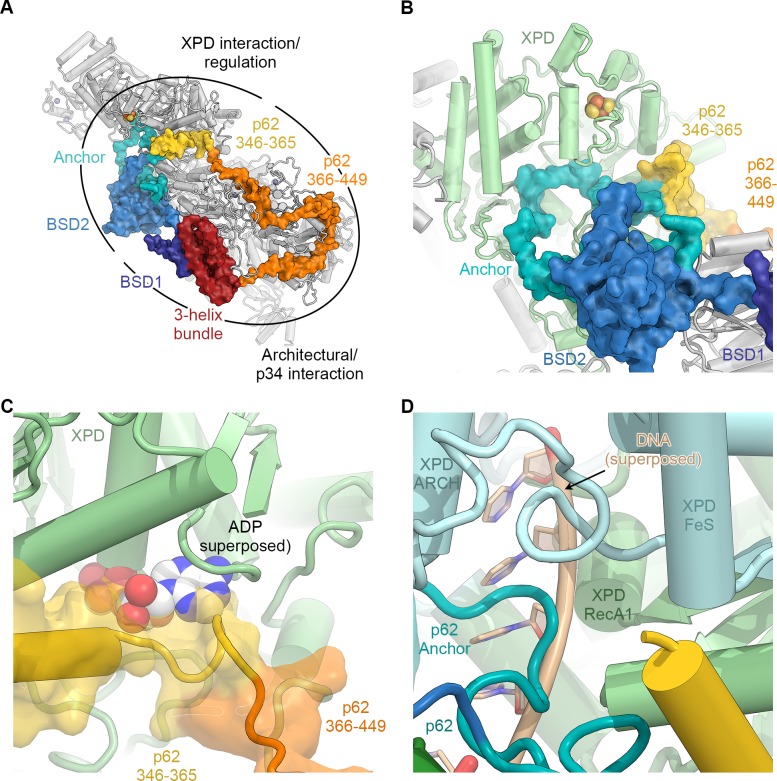
D) The p62 helix 295–317 is sandwiched between the BSD2 region of p62 and XPD RecA2, thereby bridging these two elements of TFIIH. (
E) XPD-bound DNA was modeled by domain-wise superposition of the structure of the substrate-bound helicase DinG (
Cheng and Wigley, 2018) onto the RecA-like domains of XPD. The presence of the p62 anchor near XPD RecA2 leads to steric hindrance with the DNA that would prevent binding. (
F) Residues 346–361 of p62 block access to the XPD nucleotide-binding pocket. (
G–J) Analysis of p62 (teal) conformational dynamics. (
G) Particle images were sorted for p62 elements near XPD using signal-subtracted focused classification (mask shown in yellow; see
Figure 1—figure supplement 3). (
H) Refined class with p62 density near XPD. (
I) Refined class without p62 density near XPD. (
J) Difference density (red) between the densities shown in H, I. Both classes show intact particles, even in the absence of the p62 segment inserted into the XPD substrate binding cavity.