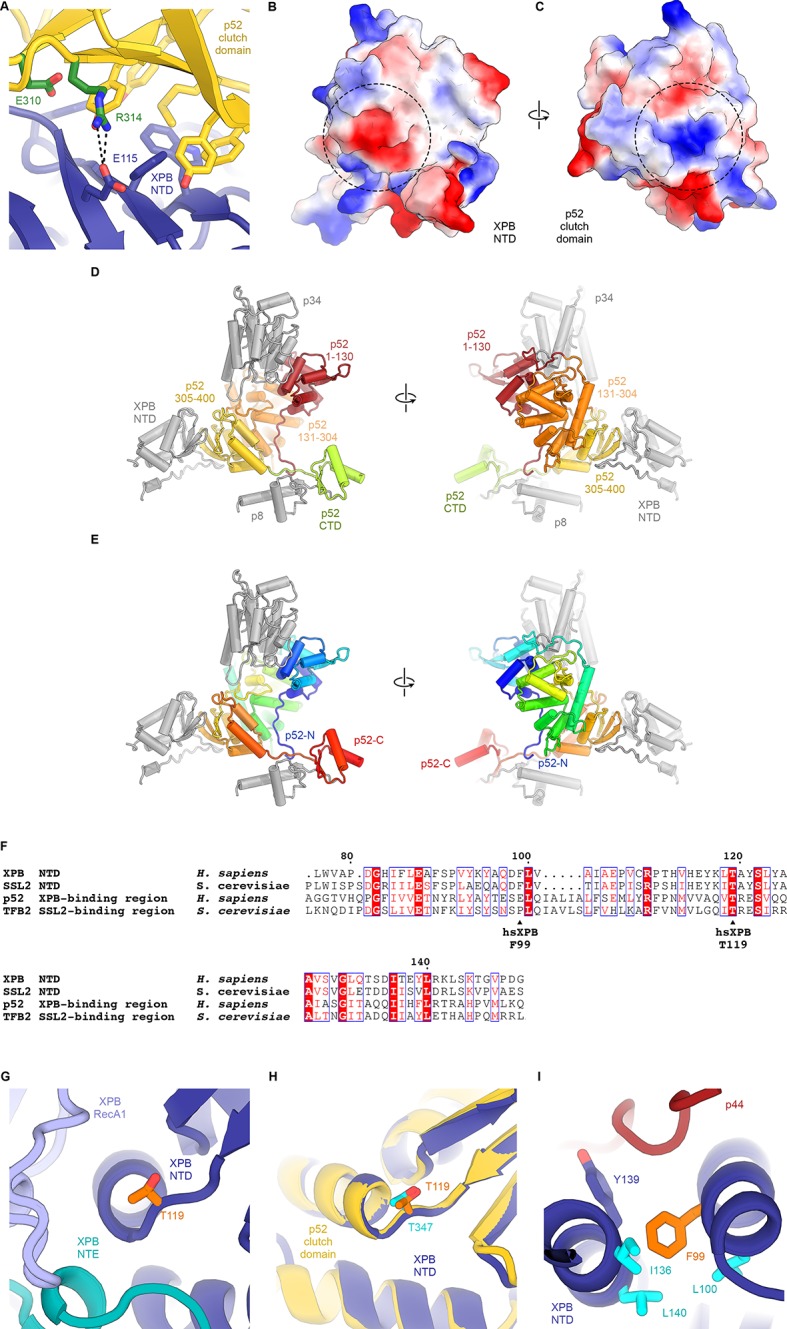
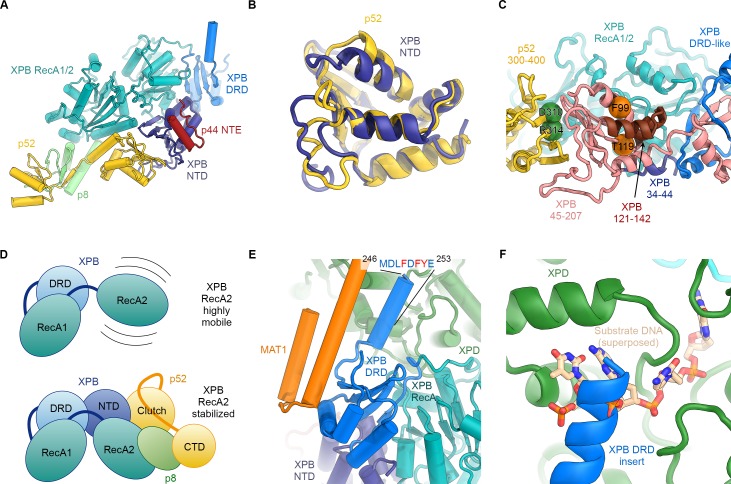
Figure 3. Structure and interactions of XPB.
(A) Bottom lobe of TFIIH. XPB RecA1/2 teal, DRD blue, NTD dark blue, p52 yellow, p8 green, p44 NTE red. (B) Superposition of the XPB NTD and the p52 clutch domain. (C) Mapping of mutations on the XPB NTD and the p52 clutch domain; mutated regions are color-coded or shown as spheres (see text for details). (D) The combined interactions of the p52 clutch, the p8-p52 CTD dimer, and the XPB NTD with XPB RecA2 may restrict the conformational flexibility of XPB RecA2 to optimize XPB activity. (E) An extension of the DRD (blue) contacts XPD (green). The sequence for which formation of an α-helix is predicted (Kelley et al., 2015) is indicated. (F) The DRD extension overlaps with the substrate-binding site on XPD RecA2. Substrate DNA modeled from PDB ID 5HW8 (Constantinescu-Aruxandei et al., 2016).
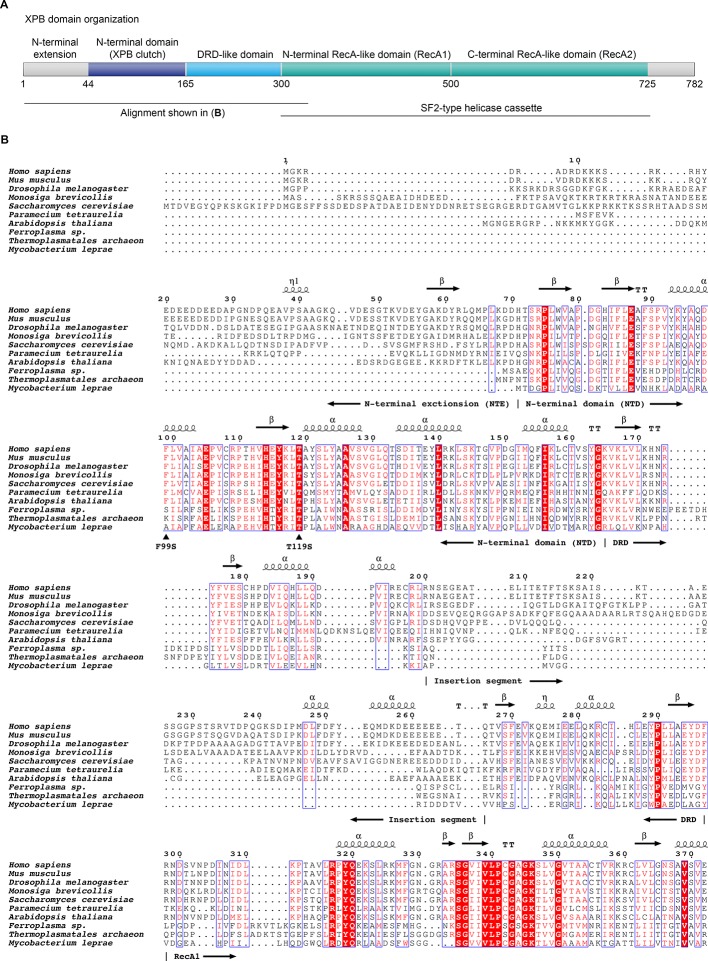
Figure 3—figure supplement 1. Domain organization of XPB and sequence alignment of the N-terminal regions of XPB or XPB-like enzymes from eukaryotes, archaea, and bacteria.
Figure 3—figure supplement 2. Structure and interactions of the XPB DRD and NTD.