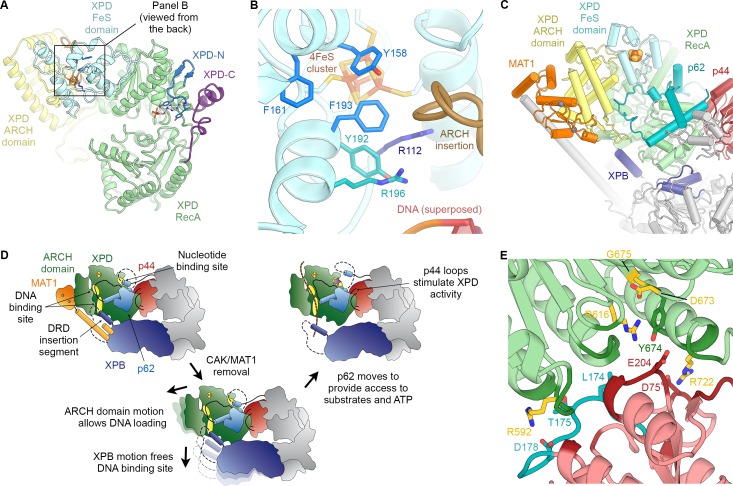
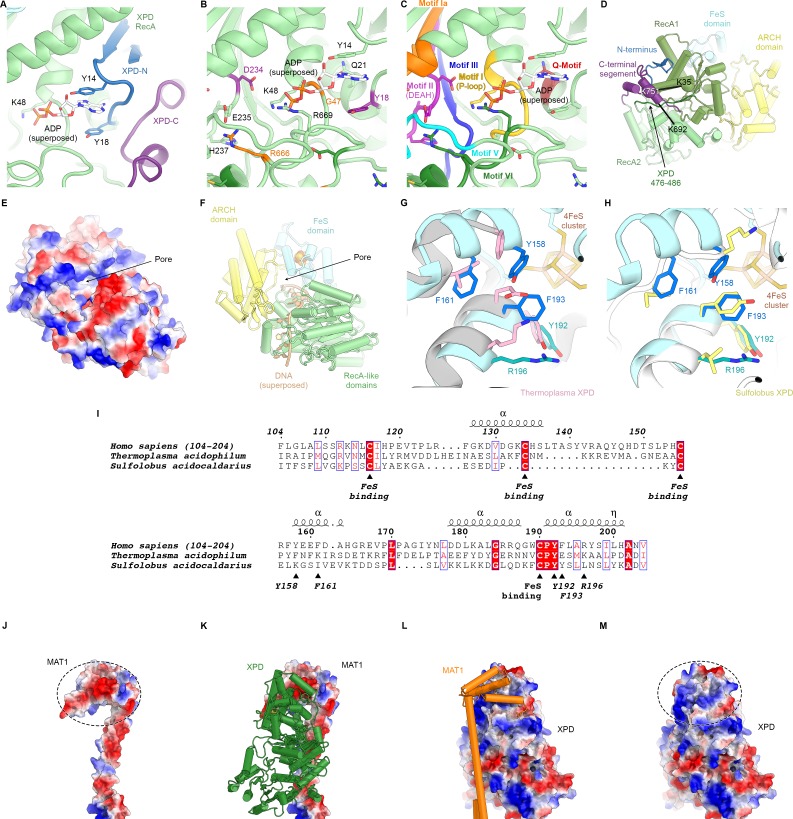
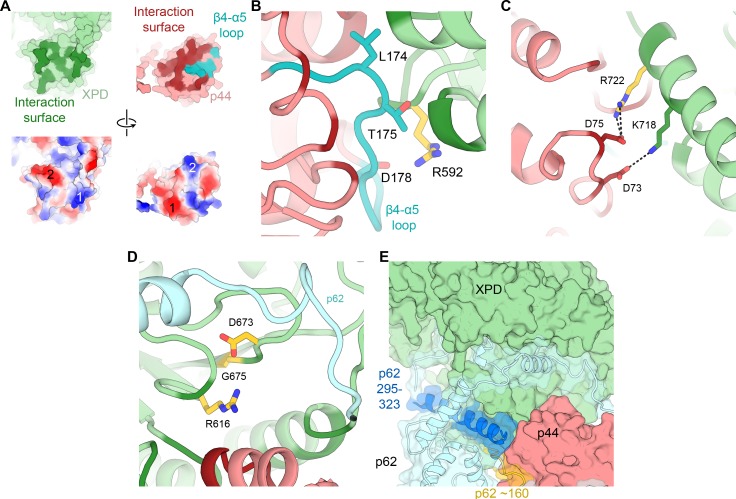
Figure 5. Structure and regulation of XPD.
(A) Structure of XPD colored by domain. N- and C-terminal segments (blue and purple, respectively) of XPD are indicated. An ADP molecule superposed from the structure of DinG (Cheng and Wigley, 2018) denotes the nucleotide-binding pocket in XPD RecA1, which is empty in our structure. (B) Structure of the FeS domain. Residues critical for XPD enzymatic activity (blue) and damage verification (teal) are indicated. The R112H mutation causes TTD in human patients. ARCH domain insertion brown. DNA superposed from (Cheng and Wigley, 2018). The region corresponding to the view in this panel (but viewed from the back side) is indicated in (A). (C) Interaction network of XPD with surrounding TFIIH subunits (interacting regions colored, remainder grey). (D) Cartoon model for repression and de-repression of XPD by MAT1, XPB, and p62. (E) XPD-p44 interacting regions (defined as residues within <4 Å of the neighboring protein) are colored in dark green (XPD) and dark red (p44). Residues discussed in the text are shown as sticks; those with mutation data (natural variants or experimental constructs) are colored yellow on XPD, teal on p44. The remainder of the β4-α5 loop harboring the synthetic p44 mutations is colored teal as well.