Figure 4.
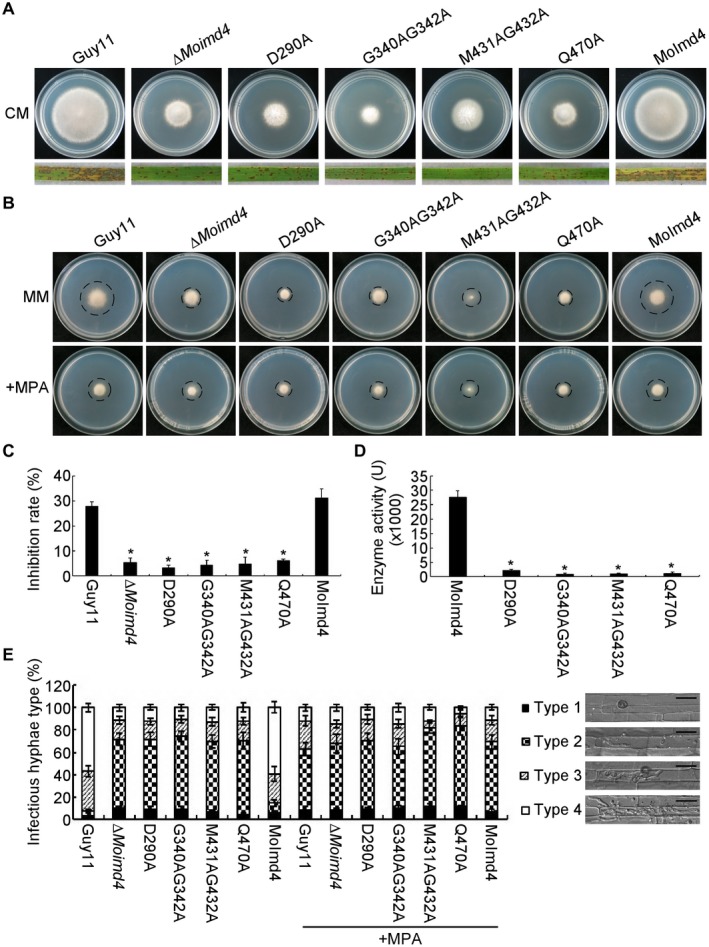
Inactivation of mycophenolic acid (MPA) binding sites leads to attenuation of MoImd4 activity. (A) Vegetative growth of Guy11, ∆Moimd4, D290A, G340AG342A, M431AG432A and Q470A mutants, and the complemented strains, on complete medium (CM) at 7 days in the dark. Conidial suspensions (5 × 104 spores/mL) of the indicated strains were sprayed onto 11‐day‐old rice seedlings. Photographs were taken at 7 days post‐inoculation (dpi). (B) Vegetative growth of Guy11, ∆Moimd4, D290A, G340AG342A, M431AG432A, Q470A and the complemented strains on minimal medium (MM) treated with or without 10 µg/mL MPA. (C) Inhibition rates of the indicated strains on MM treated with or without 10 µg/mL MPA. Experiments were repeated three times with similar results. The error bars indicate the standard deviation of three replicates. Asterisks indicate statistically significant differences (Duncan’s new multiple range test, P < 0.01). (D) Detection of enzymatic activities of the indicated strains in vitro. Target proteins were expressed in Escherichia coli BL21‐CodonPlus (DE3) cells. We defined the production of 1 mM xanthosine monophosphate (XMP) per milligram of protein per minute as 1 U of enzyme activity. Experiments were repeated three times with similar results. The error bars indicate the standard deviations of three replicates. Asterisks indicate statistically significant differences (Duncan’s new multiple range test, P < 0.01). (E) Statistical analysis of IHs of the indicated strains with or without 10 µg/mL MPA at 48 h post‐inoculation (hpi); approximately 100 IHs were counted and the experiments were repeated three times. The error bars indicate the standard deviations of three replicates. Asterisks indicate statistically significant differences (Duncan’s new multiple range test, P < 0.01). The four types of grading standard are shown, as in Fig. 3C. Bar, 10 μm.
