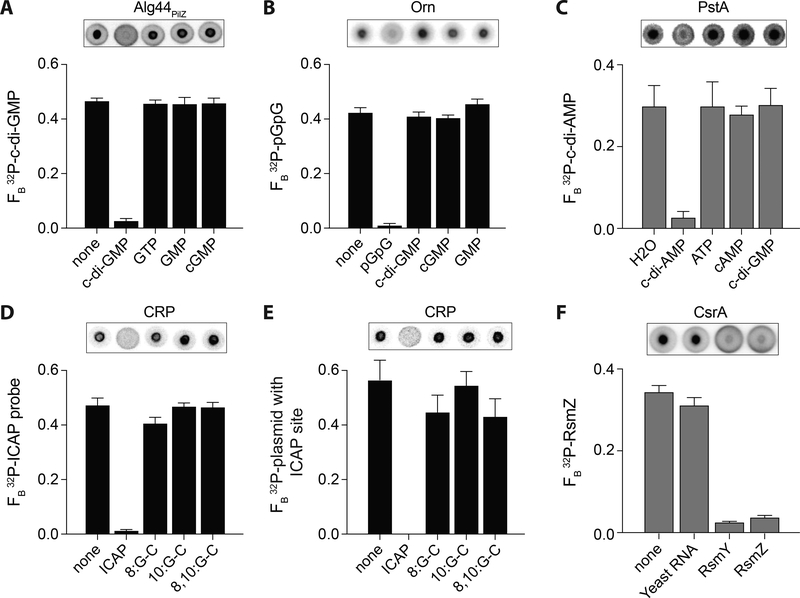
Figure 4.
DRaCALA detection of specific protein-nucleotide, protein-DNA, and protein-RNA interactions.
DRaCALA allows detection of specific protein-nucleotide, protein-DNA, and protein-RNA interactions in purified proteins (black bars) and overexpressed Escherichia coli T7Iq whole cell lysates (gray bars). Representative images of DRaCALA spots are shown above the graph. Competitors are indicated on the x-axes. (A) Binding of 32P-c-di-GMP by 2.5 μM His-MBP-Alg44PilZ in the presence of 500 μM of indicated unlabeled competitors. Adapted with permission from Roelofs et al. (2011). (B) Binding of 32P-pGpG by His-MBP-Orn (P. aeruginosa) in the presence of 250 μM of indicated unlabeled competitors. Adapted with permission from Orr et al. (2015). (C) Binding of 32P-c-di-AMP by E. coli T7Iq lysates overexpressing His-PstA (Staphylococcus aureus) in the presence of 400 μM of indicated unlabeled competitors. Adapted with permission from Corrigan et al. (2013). (D, E) Binding of 100 nM CRP with 200 μM cAMP to (D) 32P-labeled annealed oligonucleotide containing the ICAP sequence or (E) 32P-labeled plasmid with 1x ICAP site in the presence of the indicated unlabeled ICAP consensus probes or G-C substitutions at indicated positions at 1000-fold excess. Adapted with permission from Donaldson et al. (2011). (F) Binding of 32P-labeled RsmZ small RNA by E. coli T7Iq lysates (diluted 1:8) overexpressing His-CsrA (V. cholerae) in the presence of 10 μg/mL total Yeast RNA (Saccharomyces cerevisiae) or 400 nM unlabeled small RNAs RsmY and RsmZ (P. aeruginosa) competitors. Taken with permission from Patel et al. (2015).