Figure 1.
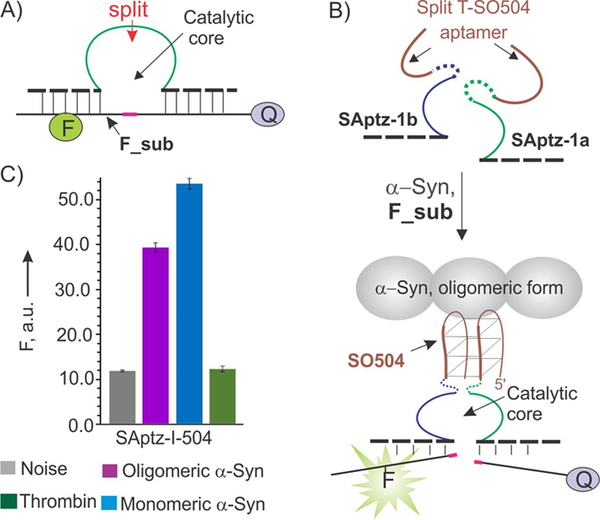
Design of split aptazyme (SAptaz) sensor for the detection of α-synuclein (α-syn). A) A catalytic DNA oligonucleotide, DZ-10–23, can cleave a fluorophore- and a quencher-labeled fluorogenic substrate (F_sub). A position for splitting the DZ as discovered by Mokany et al.[10] is indicated by the red arrow. B) Split aptazyme design strategy: both DZ and the aptamer DNA strands are split into two parts. Each half of the aptamer is linked to one-half of DZ to form SAptz-1a and SAptz-1b strands, which form the catalytic DZ core when aptameric portions bind α-syn (see Figure S1 for more details). C) Selectivity of α-syn recognition by the split SAptaz-1 sensors. All samples contained 200 nM F_sub, 2 nM SAptz-1a, 50 nM SAptz-1b, and either no protein target (Noise) or 100 nM either oligomeric or monomeric α-syn, or 26 μM of thrombin (Negative control).