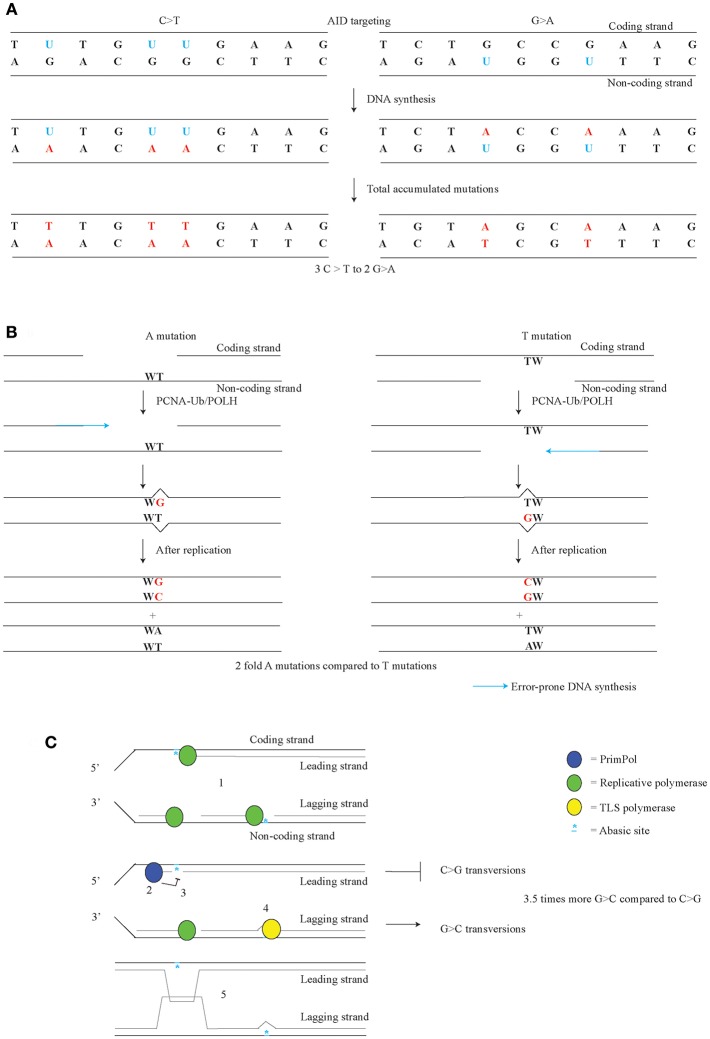
Figure 3.
Strand-biases in SHM. (A) AID targeting with a preference for the coding strand leads to a C/G transition strand bias. Us on the coding strand lead to C>T transitions, while Us on the non-coding strand lead to a G>A transitions. (B) During error-prone mismatch repair, the MSH2/MSH6 complex recognizes the U-G mismatch, after which APEX2 or PMS2 provide the incisions for EXO1. POLH is especially error-prone on template TW. Therefore, the orientation of the gap made by EXO1 likely governs the A/T bias. (C) Replicative forks can be stalled on both leading and lagging strand by AID dependent abasic sites (1). After priming on the lagging strand, a replicative polymerase resumes DNA synthesis. PRIMPOL establishes G>C over C>G transversion bias found in Jh4 intron of the Igh gene, likely though anti-mutagenic activity on the leading strand of replication. PRIMPOL restarts by repriming after stalled DNA synthesis (2) and prevents TLS (3). On the lagging strand, TLS opposite of the abasic site leads to G>C mutations (4). PRIMPOL activity likely activates a homology driven error-free pathway such as template switching to prevent mutagenesis (5). (C) adapted from Pilzecker et al. (73).