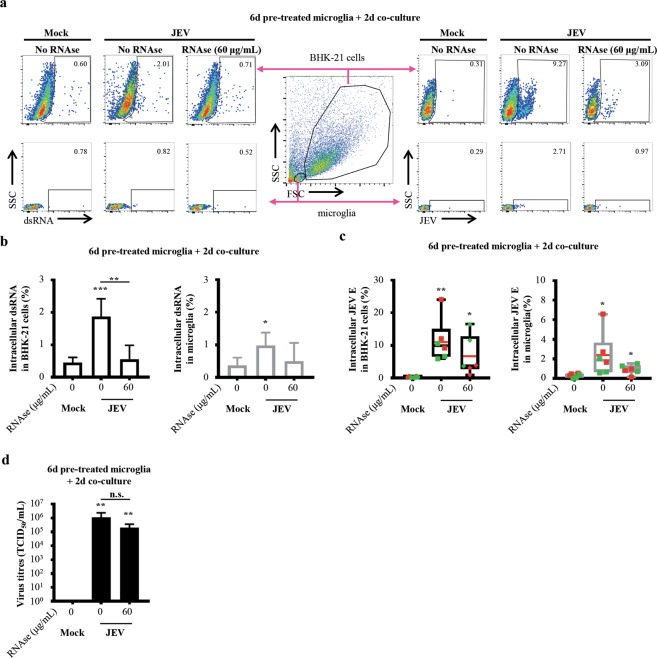
Figure 4.
Impact of RNAse A on virus transmission and recovery. Human microglia were pre-treated with Mock and JEV (Nakayama isolate used at an MOI of 10 TCID50/cell) at 37 °C for 6 days. Cells were then washed with cold PBS and subsequently co-cultured with BHK-21 cells for 2 additional days in the presence of 1:100 Ctrl serum and indicated concentrations of RNAse A. (a) Central panel is a representative density plots of flow cytometry analysis showing the gating strategy for the identification of BHK-21 cells and microglia based on their FSC/SSC profile. Other density plots represents (left panel) intracellular dsRNA and (right panel) JEV E expression in gated (upper panel) BHK-21cells and (lower panel) microglia. Frequency of dsRNA and JEV E+ cells is indicated. (b) Histogram bars representing frequencies of BHK-21 cells and microglia expressing dsRNA identified as in (a). The bar represents the mean value; the error bars the standard deviation. (c) Box plots representing frequencies of BHK-21 cells and microglia expressing JEV E identified as in (a). The symbols represent replicates where each colour is an individual donor. The black line represents the median value, the red line the mean value and the error bars the standard deviation. (d) Histogram bars representing virus titres in supernatants. The bar represents the mean value; the error bars the standard deviation. Data are of 2 independent experiments with each condition performed in duplicate or triplicate cultures. Asterisks on top of a condition show significant differences compared to mock; asterisks on black line show significant differences between the indicated conditions. Statistics are calculated with (b,c) the t-test or (d) the Mann-Whitney test (n.s.: not significant; *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001).