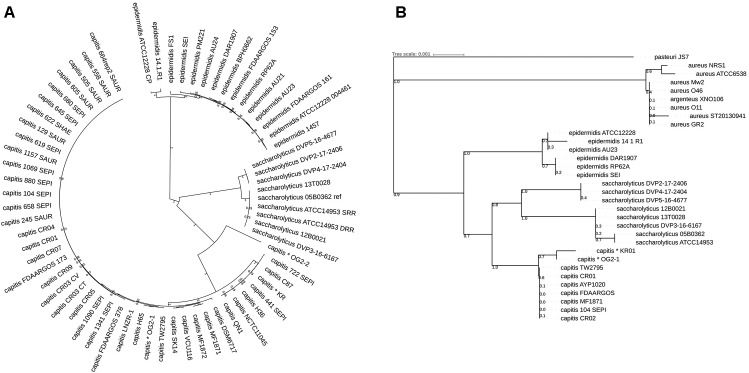
FIGURE 1.
Phylogenetic relationship of staphylococcal species based on core genome-located SNPs and 16S rRNA gene analysis. (A) Phylogenic analysis was based on high-quality SNPs in the staphylococcal core genome of all so far sequenced strains of the species Staphylococcus saccharolyticus, Staphylococcus capitis, and selected strains of Staphylococcus epidermidis, using the program Parsnp. All S. saccharolyticus strains formed a distinct clade that is clearly distinct from S. capitis and S. epidermidis. The core genome of the shown strains is 3% (70.5 kb) of the reference genome S. saccharolyticus strain 05B0362. The choice of the reference genome had no influence on the outcome of the phylogenetic analysis. The S. capitis strains that were previously incorrectly assigned as S. saccharolyticus are marked by an asterisk. (B) A Blast search with the 16S rRNA gene sequence of S. saccharolyticus strain 05B0362 was carried out and the closest matching sequences from other staphylococcal species were extracted and used for phylogenetic reconstruction. Shown are only the strains from which a complete 16S rRNA gene could be retrieved. The evolutionary history was inferred using the Minimum Evolution method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. Evolutionary analyses were conducted in MEGA7.